Perfect navigation: How to fit a compass needle into a helical bacterium?
Published in Microbiology

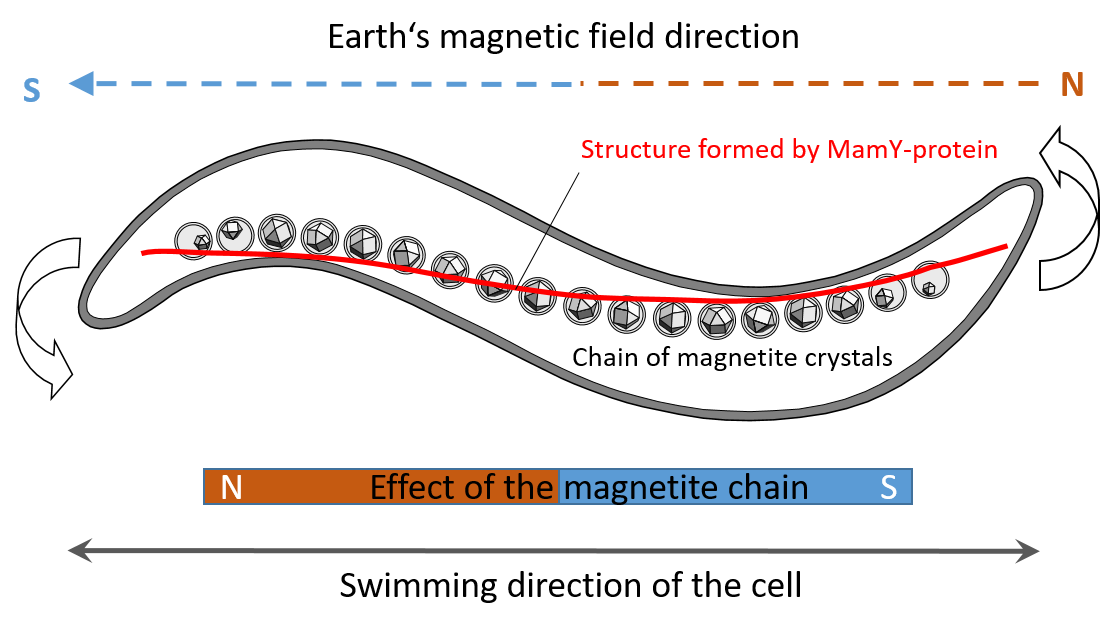
Honeybees, migratory birds, sharks, turtles and many more animals take advantage of the Earth’s magnetic field as omnipresent signpost to find distant targets. However, despite many and sophisticated attempts to understand animal magnetotaxis, the molecular base for this enigmatic sense remains elusive. By comparison, magnetotactic bacteria represent a presumably more simple, single-celled system in which magnetotaxis is much more definite. For example, it has long been known that cells of the bacterium Magnetospirillum gryphiswaldense are magnetotactic by their up to 50 intracellular magnetosomes, magnetite nanocrystals surrounded by a membrane, which are attached to a cell-spanning filament made of the actin-like MamK protein. This attachment causes the magnetite crystals not to clump together as a result of their own magnetic pull, but to become lined up, thus assuming the function of a compass needle. This enables the bacteria to follow the orientation of the Earth's magnetic field during their swimming movements and thus reach their preferred habitat, the sediments of natural waters, more quickly.
The discovery of the aktin-like filament (and the associated protein MamJ) several years ago seemed to answer all major questions about construction of a “compass needle” and performing magnetotaxis in bacteria. However, especially in magnetotactic spirilla, some intriguing issues remained: It was a mystery why this intrinsically flexible chain of magnetosomes took such a stable, linear form – while the bacterial cell body comes close to a corkscrew, i.e., there was no straight cellular structure obvious which could support the linear track of the magnetic organelles. Moreover, short fragmented magnetosome chains had been observed in mutant strains that lack the actin-like MamK and hence any discernible stabilizing element. These puzzling observations led to the hypothesis that there has to be another supporting protein that would help magnetotactic bacteria create their straight compass needle. Yet, such protein could not be identified by many years.
The answer came by screening a set of newly made deletion mutants devoid of putative genes for magnetosome biosynthesis. Among them was a strain with deleted mamY, a gene so far implicated in other functions such as magnetosome vesicle formation in a related magnetotactic bacterium. Looking at the TEM images, we thus got disappointed, as the cells seemed to contain regular chains of magnetosomes. Nevertheless, the chains appeared somehow unfamiliar: They were bent and localized to the "wrong" side of the cell body. To understand that we may have hit here a missing chain-localizing factor took us some time as this required to mentally convert the two-dimensional TEM micrographs from Magnetospirillum cells back into three-dimensional helices with an internal magnetosome chain. This insight was the starting point for an in-depth analysis of MamY, which finally turned out to be a key-player of an appealing and unexpected concept of how helical magnetotactic bacteria embed a straight “compass needle” in their spiral shaped cell body.
Altogether, we now can define a sophisticated “magnetoskeleton” which to date consists of three cytoskeletal factors dedicated for magnetotaxis. The actin-like MamK and its adaptor MamJ represent the dynamic part of the magnetoskeleton as they actively concatenate magnetosomes and position the chain at midcell, thereby also ensuring precise splitting and equipartitioning of the magnetoreceptor upon cell division. The static MamY structure by contrast is responsible to identify and to tether the dynamic assembly to the geodetic cell axis, i.e. to fix it along the shortest path connecting the cell poles. This straightens the “compass needle” within the helix and aligns it with the motility axis, thereby perfectly superimposing the Earth's magnetic field vector on swimming movements. These results highlight that the phenomenon of bacterial magnetotaxis still bears unexpected and sophisticated features waiting for discovery.
Check out the video:
and read the full paper at https://www.nature.com/articles/s41564-019-0512-8
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in