Protein abundance of a bacterial predator uncovers a bacteria-damaging enzyme
Published in Microbiology and Cell & Molecular Biology
Worldwide, antibiotic resistant bacteria are responsible for approximately one in six confirmed bacterial infections in humans (2023)1. Despite antibiotic stewardship measures, these alarming numbers continue to rise and are indicative of the urgent need for new ways to fight antibiotic resistant bacterial pathogens. Luckily, in nature a variety of different predators of bacteria exist. While undoubtedly, the most well-known are viruses of bacteria (bacteriophages), which are used in phage therapy, there are also predatory bacteria that kill other bacteria. Amongst these, Bdellovibrio bacterivorous has the most promising properties as a potential ‘living antibiotic’. Bdello, what you say? While indeed the name of this tiny predator is barely pronounceable, we already know a lot about it and procedures have been developed to formulate this bacterium for use against other bacteria2,3. It is known that B. bacteriovorus kills a wide range of other bacteria including those resistant to antibiotics4,5. B. bacteriovorus kills, invades and digests the bacterial prey before damaging and exiting the prey remnants in the end. My goal is to ensure more people know about this fascinating predatory bacterium and what this bacterium is doing to make the most of its properties in the future.
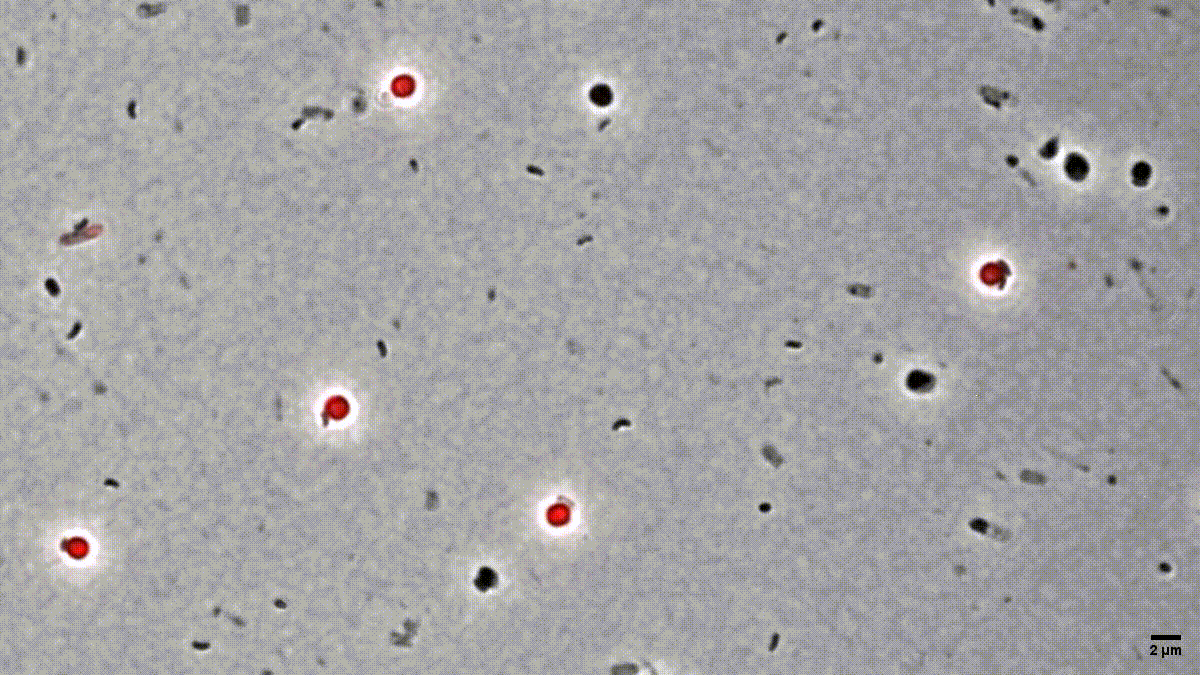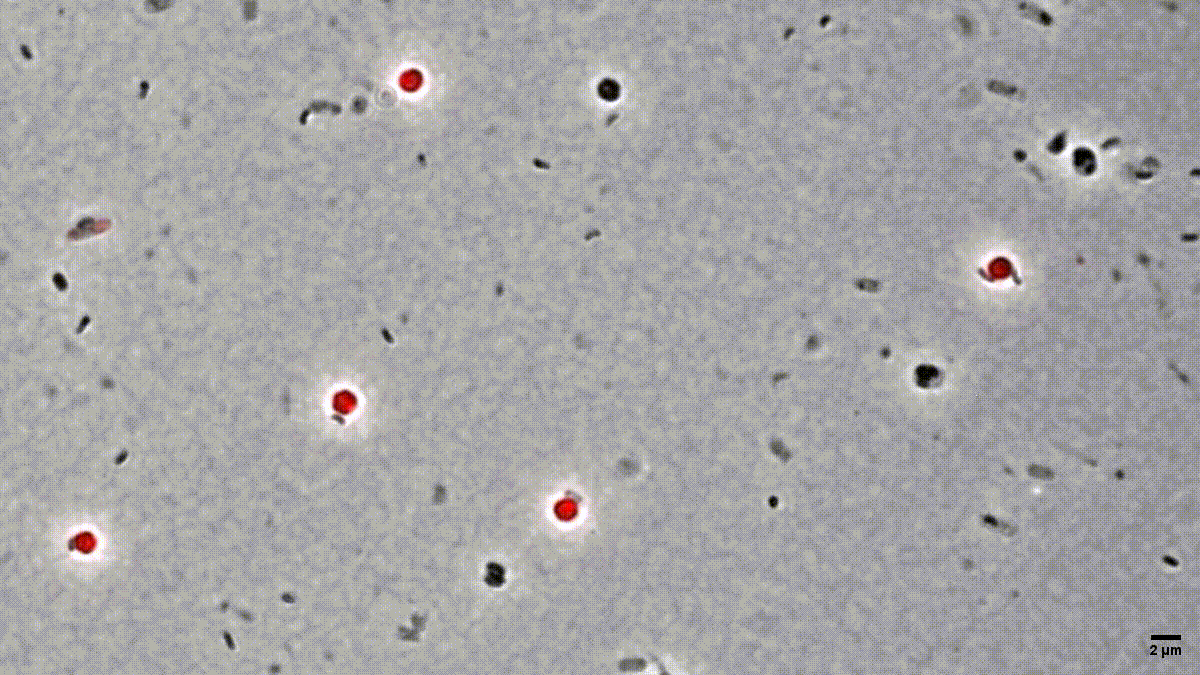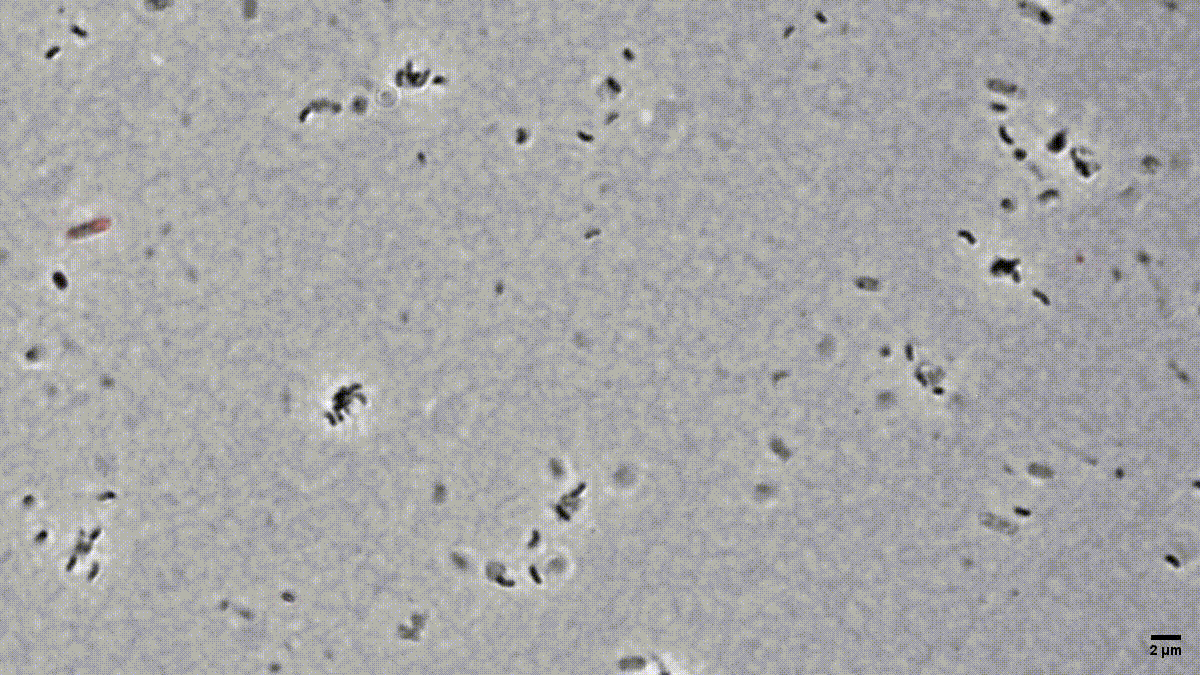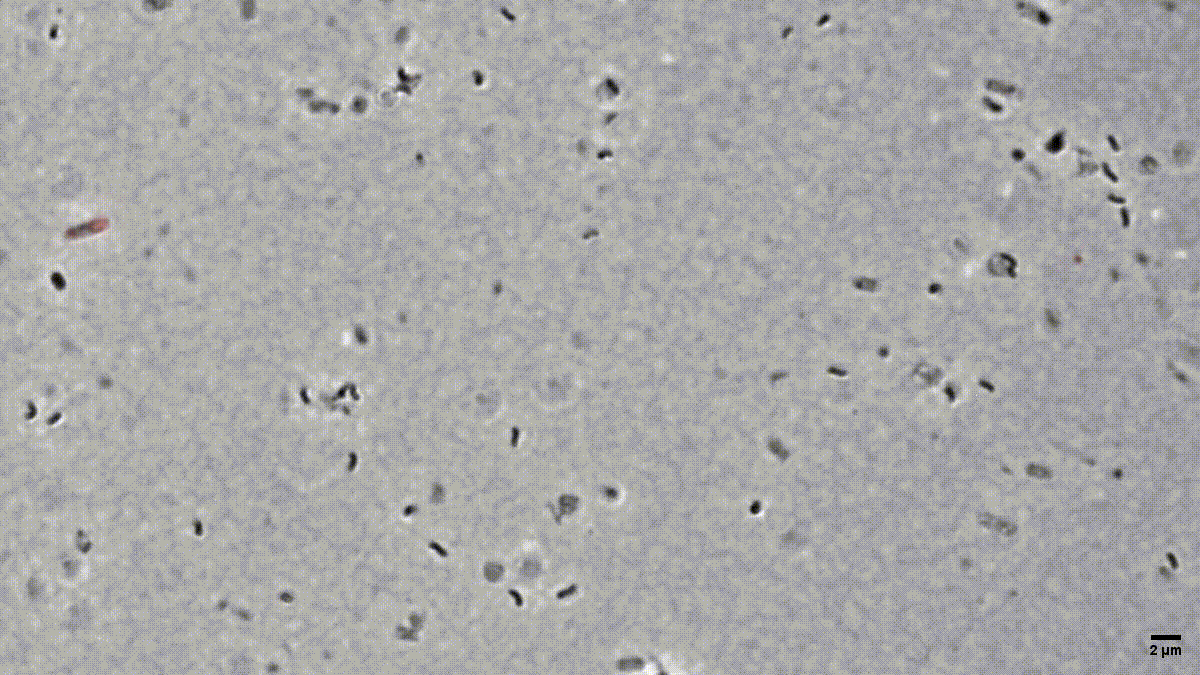
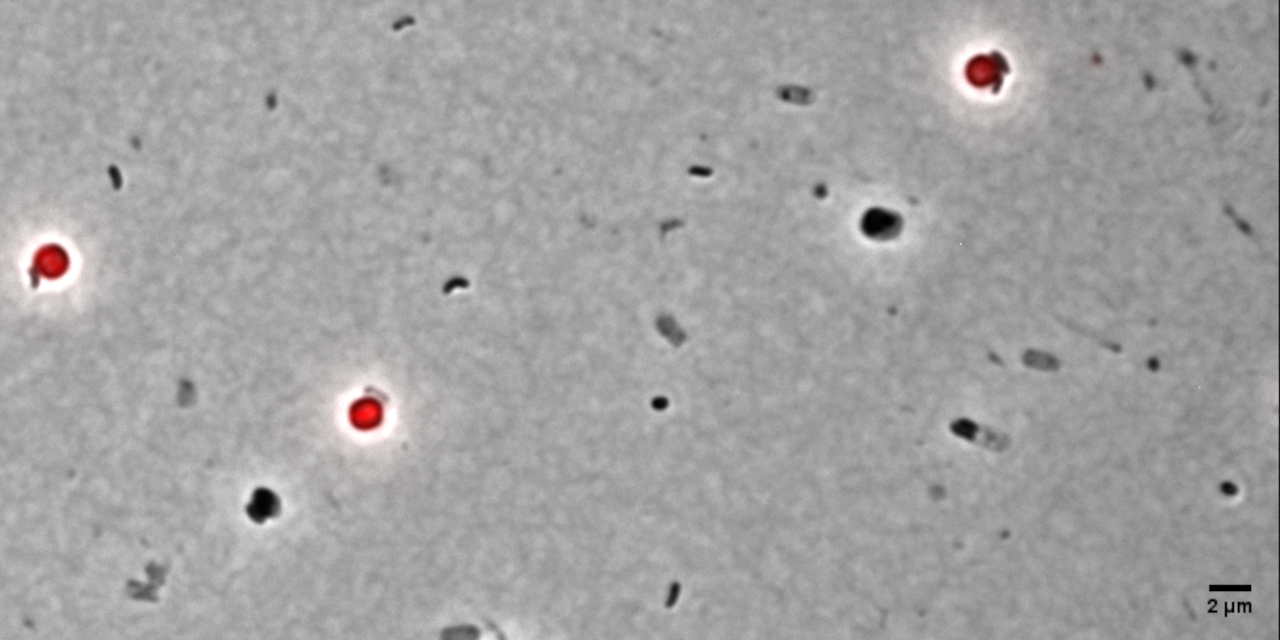
Figure 1: Time-lapse microscopy of multiple small B. bacteriovorus cells lysing and exiting the prey remnants (E. coli prey cytosol in red). Movie recorded by D. Jankov, processed by S. Huwiler.
Many people are fascinated when they see how Bdellovibrio break down, or lyse, Gram-negative bacterial pathogens after it has eaten up the prey from the inside (Fig. 1). But what is actually happening? How does predatory B. bacteriovorus manage to get out of its bacterial prey remnants? Measuring RNA levels is a good basis to understand the molecular mechanisms involved, but in the end, the critical cellular functions that achieve bacterial damage and opening up of the prey cell remnants are mainly performed by proteins. This is why we set out to generate the first quantitative predatory bacterium proteome over the entire predatory life cycle from the point at which B. bacteriovorus enters the periplasm of a model prey bacterium, Escherichia coli, during internal development until the predator kills and breaks out of its prey remnants. Specifically, Ting Lai generated and processed samples from five biological replicates over the entire predation cycle. Then, with help from the Functional Genomics Center Zurich, we used these samples to produce a highly resolved and quantitative pattern of protein abundances in B. bacteriovorus (Fig. 2). Revealing which proteins from the predator are how abundant at different stages during the predation process, could provide us hints to their relative activity at each stage. In conjunction with predicted protein structural information and homology, this in turn can provide us critical information about what each protein is actually doing. Of particular interest to us are predator proteins, particularly enzymes, that damage bacterial prey through cell lysis, and are specifically active when B. bacteriovorus is exiting the prey remnants.
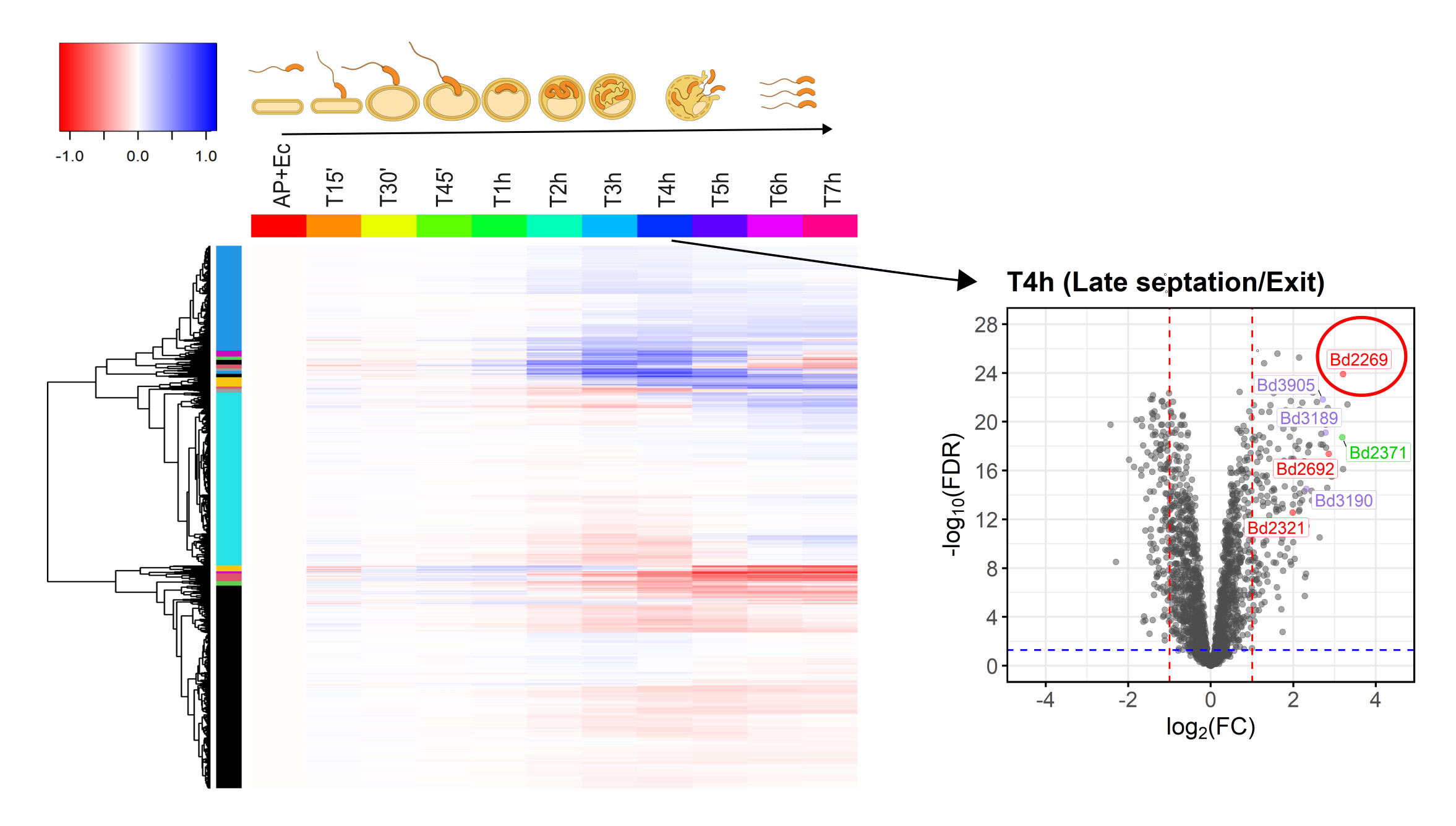
Figure 2: Heatmap overview of B. bacteriovorus proteins identified by quantitative proteomics over the entire predatory life cycle (T15’-T7h) in E. coli prey (highly abundant in blue, low abundance in red). A volcano plot of this dataset (on the right) identifies predatory protease Bd2269 as up regulated in the prey exit phase (red encircled). In the heatmap protein abundances were normalized to the initial condition, where B. bacteriovorus is free swimming (AP = attack phase) and the E. coli is still on its own (Ec).
Therefore, we did not stop where many ‘omics’ studies end at the descriptive part, but continued to attribute a function to a protein that was abundant when B. bacteriovorus was leaving the prey remnants, specifically the enzyme, protease Bd2269. By knocking out the gene responsible for production of this enzyme, we showed that, indeed, this protease was involved in mediating the exit process of B. bacteriovorus from prey remnants. Furthermore, when Bd2269 was expressed within the E. coli prey, where it is not normally produced, it lysed the E. coli from within. Because this enzyme normally acts inside the gram-negative bacterial prey, more research will be needed to use this enzyme practically as an external application for bacterial pathogen control. Despite this, the same enzyme is known to actively reduce Staphylococcus aureus biofilms6, so it has great potential.
This quantitative proteome provides the research community with a valuable resource for studying critical stages of the predatory process and advances the search for novel antimicrobial enzymes. So, next time you hear about microbial warfare or ‘helpful’ bacteria, I would be delighted if you remember Bdellovibrio, a fascinating bacterium with a tongue twisting name that is a treasure trove for understanding antimicrobial mechanisms and has great potential as a ‘living antibiotic’.
This paper is available at: https://www.nature.com/articles/s42003-025-09010-x
References:
- World Health Organization. Global Antibiotic Resistance Surveillance Report 2025. https://www.who.int/publications/i/item/9789240116337 (2025).
- Cao, H., Wang, H., Yu, J., An, J. & Chen, J. Encapsulated Bdellovibrio powder as a potential bio-sisinfectant against whiteleg shrimp-pathogenic vibrios. Microorganisms 7, 244 (2019).
- Sason, G. et al. Encapsulated predatory bacteria efficiently protect potato tubers from soft rot disease. Plant Disease https://doi.org/10.1094/PDIS-02-24-0487-RE (2024) doi:10.1094/PDIS-02-24-0487-RE.
- Sun, Y. et al. Predation Efficacy of Bdellovibrio bacteriovorus on multidrug-resistant clinical pathogens and their corresponding biofilms. Jpn J Infect Dis 70, 485–489 (2017).
- Upatissa, S., Mun, W. & Mitchell, R. J. Pairing Colicins B and E5 with Bdellovibrio bacteriovorus to eradicate Carbapenem- and Colistin-resistant strains of Escherichia coli. Microbiol Spectr 11, e00173-23 (2023).
- Im, H., Dwidar, M. & Mitchell, R. J. Bdellovibrio bacteriovorus HD100, a predator of Gram-negative bacteria, benefits energetically from Staphylococcus aureus biofilms without predation. The ISME Journal 12, 2090–2095 (2018).
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in