Rational design of ligands based on dynamic surface adsorption affinity
Improving the efficiency and stability of perovskite solar cells require a thorough examination of surface defect chemistry and physics. This understanding serves as a cornerstone for the rational molecular design of defect-passivating ligands. Prior studies have primarily designed passivating molecules based on a static picture of the interaction between passivating ligands and perovskite surfaces under vacuum at 0 K; these models do not take into account operational and atmospheric stressors.
In this work, we first employed DFT calculations (HSE+SOC method) to assess the formation energies of charged defects at both MAI-terminated and PbI2-terminated surfaces, as well as in the bulk. Notably, we observe distinct properties of surface defects compared to bulk defects. For instance, donor-like defects tend to be deeper at the MAI-terminated surface, whereas acceptor-like defects tend to be deeper at the PbI2-terminated surface.
We posited that surface passivators for mixed Pb-Sn perovskites should ideally have multi-functional units encompassing electron-rich and electron-poor domains (Fig. 1a). We reasoned that a higher maximum electrostatic potential (φmax) at the electron-poor side (NH3+/NH2) promotes stronger interactions with acceptor-like defects at the perovskite surface, while a lower minimum electrostatic potential (φmin) at the electron-rich side (SO3/SO2/PHO3) could increase the binding strength with donor-like defects.
We conducted ab initio molecular dynamics (AIMD) simulations to investigate the dynamic behavior of surface defects, and the dynamic adsorption affinity (DAA) of two passivators – 3-amino-1-propanesulfonic acid (APSA) and 4-aminobutylphosphonic acid (4-ABPA) – on the perovskite surface under device operating conditions such as exposure to oxygen, moisture and heat (Fig. 1c).
We introduce a novel approach to designing passivator structures that differs from static interaction models without temperature effects and time scales. Instead, our method prioritizes dynamic surface adsorption ratios under operational environmental conditions, offering a fresh theoretical approach to suppress degradation at the perovskite surface. We found that though APSA and 4-ABPA have a comparable static binding energy with the perovskite surface (Fig. 1b), the latter exhibited a much stronger DAA and improved stability under operational conditions.
Our findings indicate that 11 4-ABPA ligands were adsorbed on the perovskite surface through strong O-Pb (Sn) coordinate bonds. However, in the case of APSA adsorption, only 7~8 under-coordinated Pb2+/Sn2+ sites at the perovskite surface were coordinated (Fig. 1d). Moreover, during the last 2ps AIMD simulations, the number of adsorbed ligands showed a wider fluctuation range (4~10) in the case of APSA passivation, while 4-ABPA has a narrow fluctuation range of 11~12, indicating more stable surface binding.
Leveraging this DAA-guided design approach, experimental efforts yield a substantial improvement in carrier lifetime for perovskite films. The resulting narrow-bandgap devices demonstrate a tenfold enhancement in operational stability, underscoring the efficacy of our approach in advancing perovskite solar cell technology.
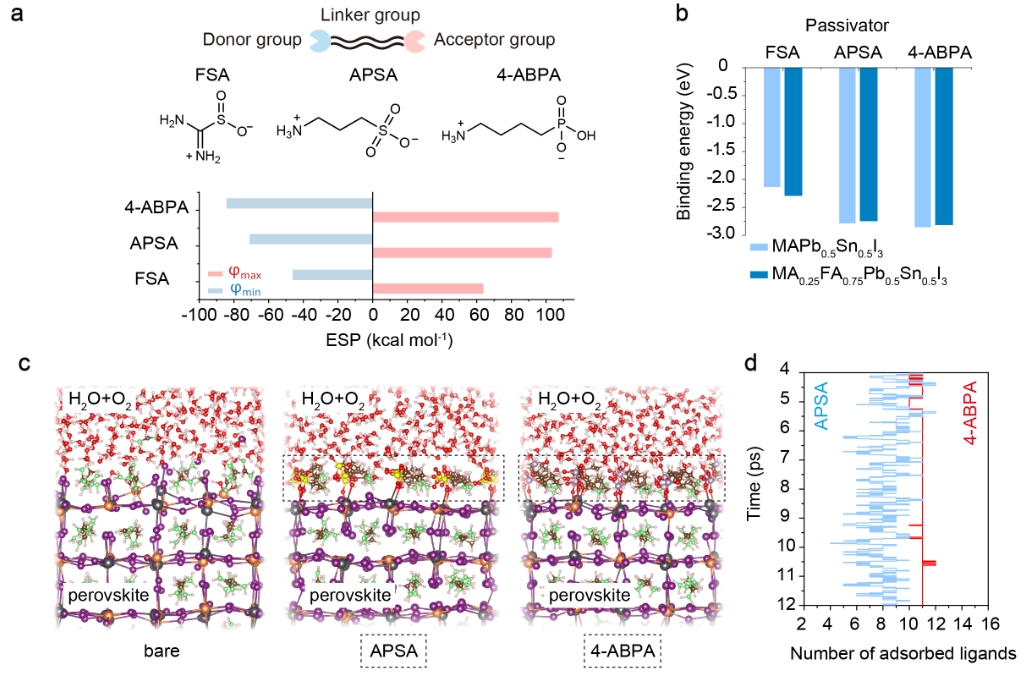
Figure 1 Design of passivators. a, Molecular structures of three passivator molecules (FSA, APSA and 4-ABPA). Gaussian calculated electrostatic potentials (φ, ESP) values of maximum ESP (φmax) and minimum ESP (φmin) are also shown. b, Binding energy of three molecules with the AI-terminated (001) surface of MAPb0.5Sn0.5I3 and MA0.25FA0.75Pb0.5Sn0.5I3. c, ab initio molecular dynamics (AIMD) snapshots of bare, APSA, and 4-ABPA adsorbed perovskite surface exposure to oxygen and moisture conditions at a temperature of 400K. The perovskite composition is MA0.25FA0.75Pb0.5Sn0.5I3. P atoms, lavender color; S atoms, yellow color; O atoms, red color; Pb atoms, grey color; Sn atoms, orange color; I atoms, purple color; N atoms, green color. d, The number of adsorbed ligands for APSA and 4-ABPA at the conditions shown in Fig. 3c from 4ps to 12ps during the AIMD simulation. Ligand number of 16 represents complete surface coverage within the simulation unit. Blue line: APSA adsorption case; Red line: 4-ABPA adsorption case. The average value of adsorbed ligands during the last 2ps of the AIMD simulation for APSA and 4-ABPA are 8 and 11, respectively.
For more details, please check out our paper “The dynamic adsorption affinity of ligands is a surrogate for the passivation of surface defects” in Nature Communications (https://doi.org/10.1038/s41467-024-46368-8).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in