Recognition of anion-water clusters by peptide-based supramolecular capsules
Published in Chemistry and Cell & Molecular Biology
The story behind our recent work on anion-water cluster recognition started with a legacy from my colleague, Dr. Alberto Fuertes. When he finished his PhD, he handed me a flask with a few milligrams of what he and our supervisor, Prof. Juan Granja, thought could be an excellent anion receptor. They figured the preorganized structure of the cyclic peptides (CP) would pair perfectly with a triazolic cap to create a suitable pocket for anions (Figure 1). Dr. Fuertes, being a wizard with tricky syntheses, managed to create the tris-triazolic peptide CP2 by coupling the alkyne-bearing CP1 with tris(2-azidoethyl)amine through alkyne-azide cycloaddition. The alternated chirality of the residues contained in CP1 sequence allow it to self-assemble though hydrogen bonds, forming dimers (D1). In the case of CP2, since it was equipped with a cap, the resultant supramolecular structure was a capsule, D2.
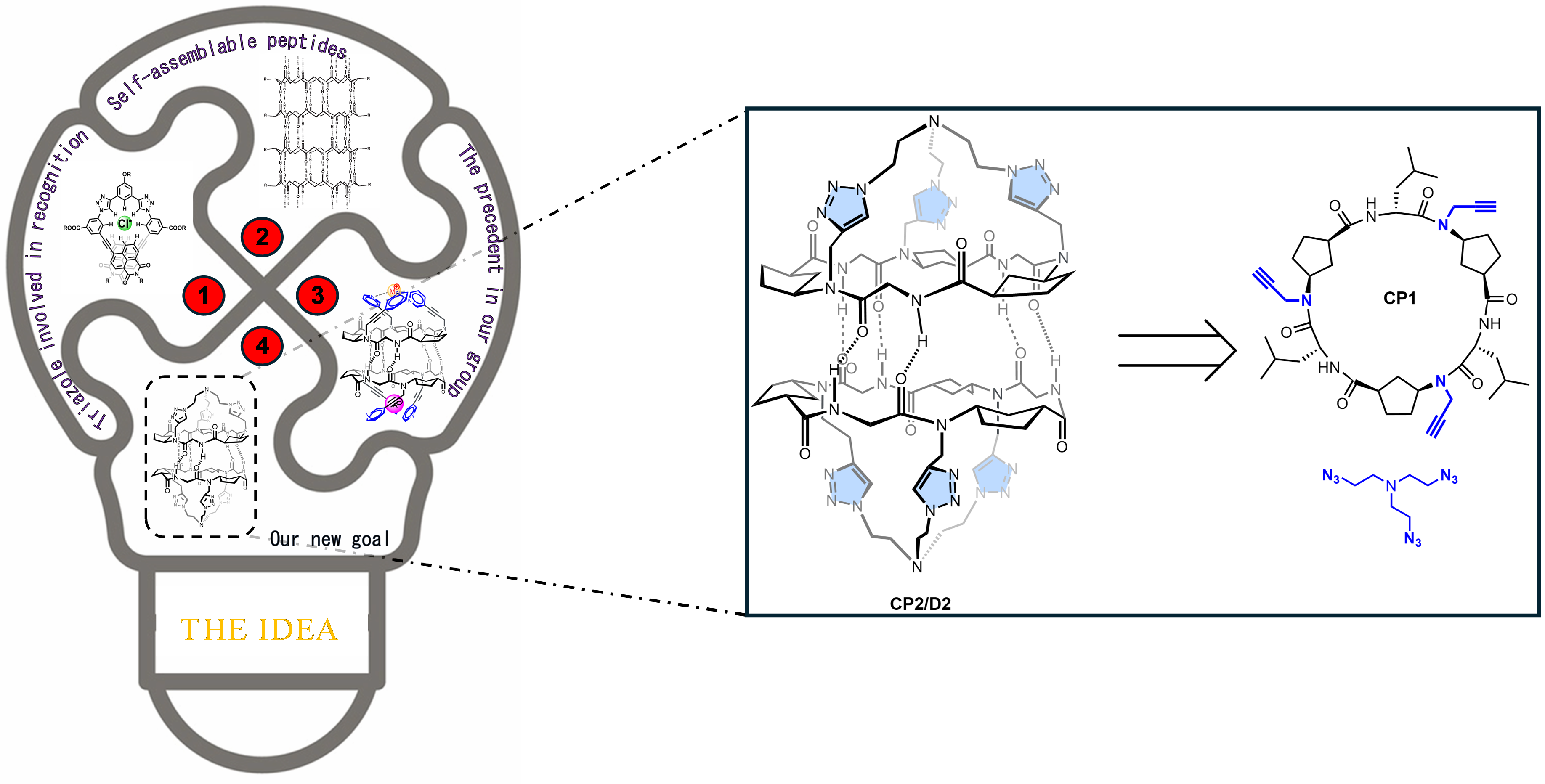
Image 1: Schematic representation of the work main idea (left) and retrosynthetic analysis of the receptor D2 from the cyclic hexapeptide CP1 and tris(2-azidoethy)amine (right).
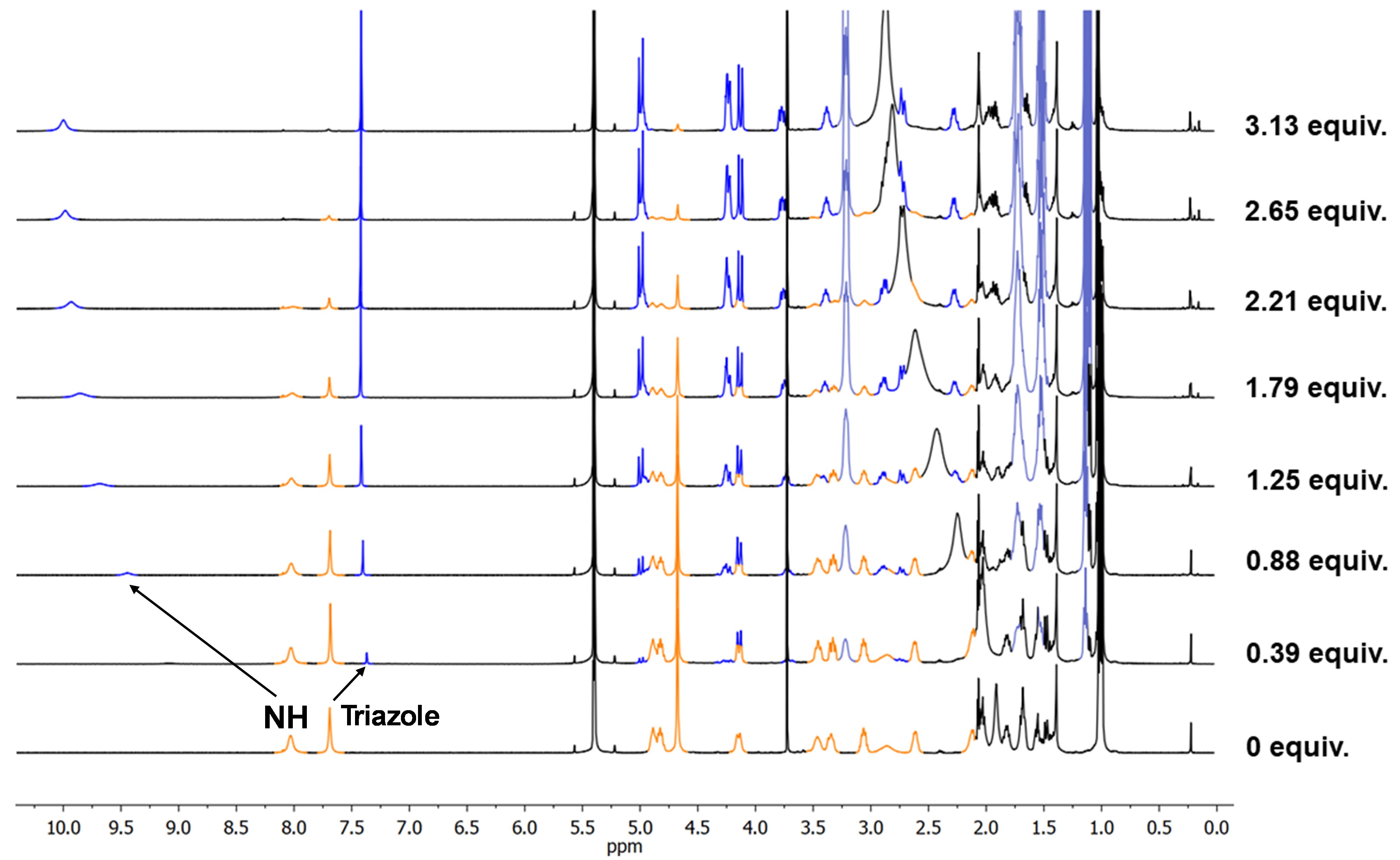
After some months in the freezer and during one of those PhD slumps where everything seems to be going wrong, I decided to give D2 a shot to brighten my PhD live. The idea was simple: some small anions could easily accomodate in the pocket created by the three triazoles of each CP2 unit. It sounded easy to check that triazole proton shift upon addition of TBAF or TBACl. A few days later, I was astonished to see the NMR monitor showing the opposite of what we expected. The triazole proton underwent upfield shift, and almost in the same extent for the two anions, fluoride and chloride. More surprisingly, the amide proton seemed to be doing something curious, being remarkably downfield shifted. My supervisor was even more shocked and almost ordered me to repeat it. Playing detective, I dug deeper and found, through nOe crosspeaks comparison, that the peptide underwent a massive rearrangement upon forming the “complexes” (even though we didn’t know what species were forming). The expression on my and my supervisor’s face, as we tried to translate the ROESY NMR data to our stick model, was priceless (see Image 2 for a rough idea).
| 1. THE OBSERVATION | 2. PROCESSING |
 |
 |
| 3. THE MODEL | |
 |
|
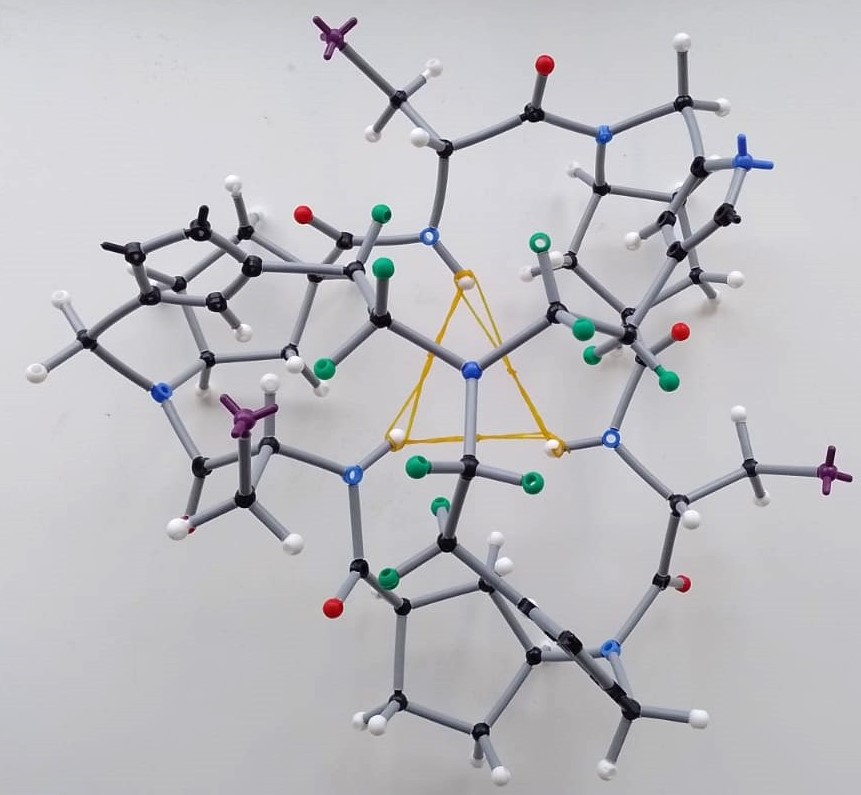
Image 2: Illustrative setps that exemplifies how bewildered we were trying to propose a plausible peptide conformation with molecular models employing the information provided by the 1D and 2D NMR studies.
We concluded that the triazole proton wasn’t directly interacting with the anion and that the new species retained tertiary symmetry. With some elastic bands to restrict the model’s mobility, we agreed the mysterious species had a triangle-like shape where the amide protons pointed sharply inside the peptide cavity, likely placing the anion around there. But a big question remained: were the complexes dimeric or monomeric? It was known in the group that these peptides (α,γ-cyclic peptides) prefer forming heterodimeric rather than homodimeric assemblies. The plan was to follow the heterodimer formation via 1H NMR and then add the anion. If the complex was a dimer, we should see the heterodimer signals shift. I prepared CP3 (which forms D3 homodimers), an analogue of CP1 but with γ-amino acids having a six-member ring (Ach) instead of a five-member one (Acp). Surprisingly, adding the anion to the heterodimer D2-3 brought back the D3 as a homodimer and formed the same complexes previously identified with D2 and F- or Cl-. This meant the heterodimer couldn’t accommodate these anions. After a second round of this experiment with D2-4 (the heterodimer between CP2 and its Ach-analogue CP4), we confirmed that peptides with the more rigid six-member ring residues couldn’t recognize the anions. This reinforced the idea that peptide rearrangement was crucial for the recognition process. Even though the triazole wasn’t actively participating, we confirmed the cap’s major role since adding TBAF or TBACl to CP1 didn’t change its 1H NMR spectra. By the way, I made those beautiful colored NMR stacked spectra with my trusty Paint.
In the middle of this research, my supervisor suggested using ITC (Isothermal Calorimetry) to get the thermodynamic parameters of the complexation process. Since I was a newbie at this, a senior colleague helped me analyze the measurements. Something was off in the spectra, and he recommended drying the TBA salt solutions completely since the hydrated salts likely affected the measure. Out of curiosity, I added some activated molecular sieves to the tube containing the complex between D2 and chloride. I didn’t know what to expect, but definitely not the recovery of the free receptor D2. I remember running into my supervisor’s office, thinking that maybe it was all due to water and not the anions…too good to be true, I thought. Fortunately, that was ruled out, and my supervisor proposed that maybe we were recognizing one hydrated anion…until we got the X-ray structures. This is a funny story. Like every chemist, my supervisor dreamed of obtaining crystals. Since it had been years since the last crystal structure reported in our group, I just assumed it was dreaming too big. I couldn’t believe it when I saw that sparkly solid (D2 at the bottom of an NMR tube) under the microscope. I sent it to the X-ray analysis unit without telling my supervisor, just in case the diffraction data weren’t good enough…but they were. I remember getting an email from the crystallographer and rushing to my supervisor’s office to see it together. Our faces spoke volumes! It was BEAUTIFUL!
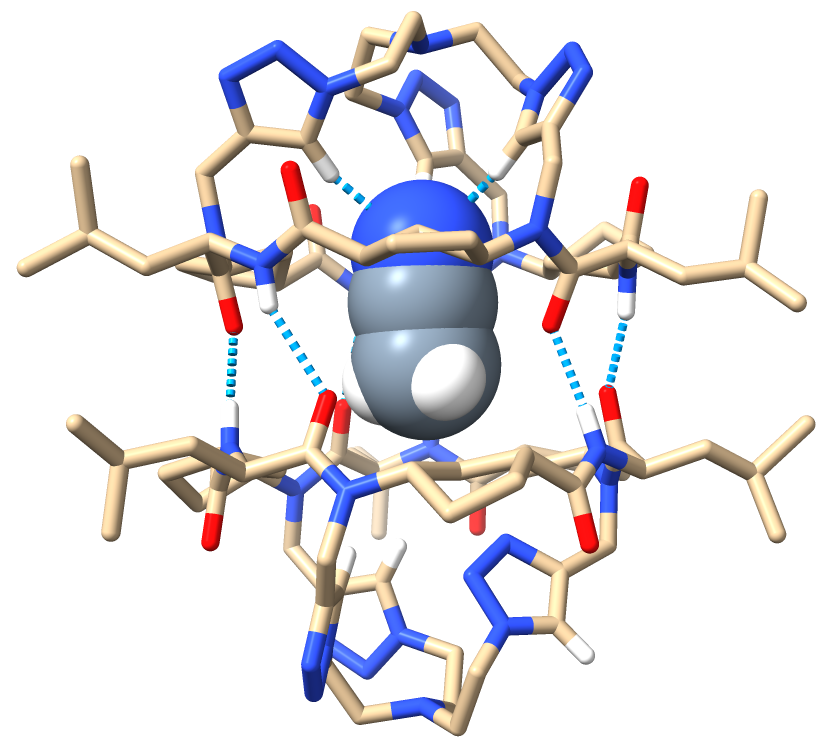
Image 3. Front view of the ACN⊂D2 crystal structure.
This was the boost that I needed to dive deeper into our system. I ran NMR titrations with other anions (I-, Br-, NO3-, N3-, AcO-, Br3- and PF6-) and discovered that the lack of response we once saw with iodide or bromide was due to their TBA+ salts having less water, not their inability to be complexed. We were excited to explore the transport activity of D2. Those weeks with my old friend, the Fluoromax-3, paid off, as we determined it ability to promote anion and proton exchange across lipid membranes.
Amidst all this, I had the opportunity to spend three months in Prof. Fahmi Himo’s group at Stockholm University. We met at a conference, and it was the perfect chance to learn about DFT calculations. Anyone who knows me knows that anything computer-related isn’t my strong suit, but there I was. I learned a lot from him and continued improving my computational skills back in Spain. Thank to that, I could polish a little bit more the research I was doing. During my stay abroad, I received an email from Dr. Antonio L. Llamas-Saiz, notifying me that he found good material in the samples I gave him before leaving, the ones containing the complexes between D2 and chloride and fluoride anions. That was the icing on the cake. Two fascinating complexes where a cluster composed by three anions and a variable amount of water molecules is embedded in between two CP2 subunits, with are not any more forming a proper dimmer.
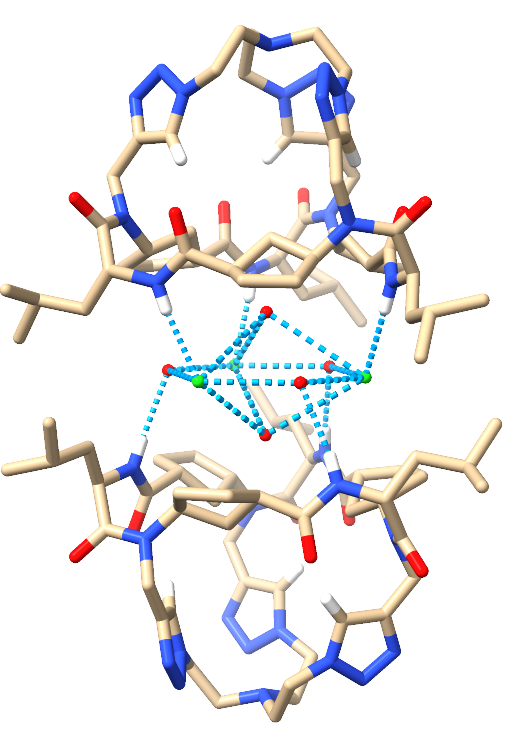 |
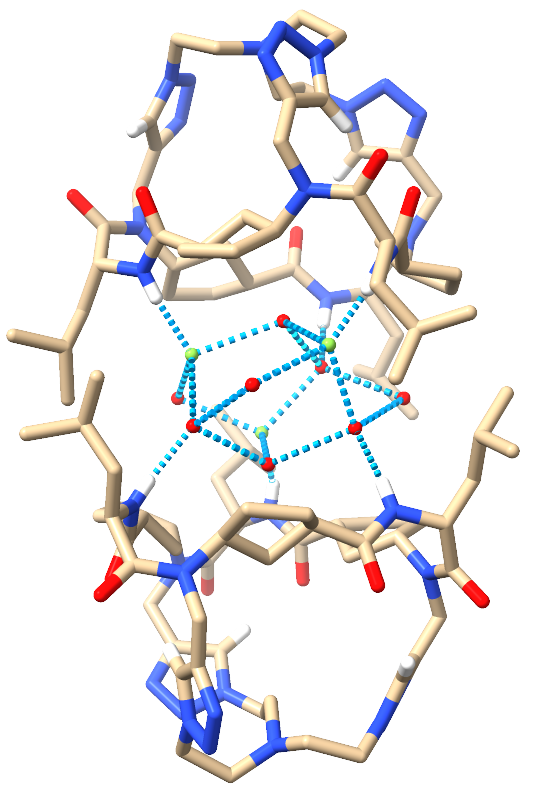 |
Image 4: Front views of the 3Cl-·4H2O⊂2CP2 (left) and the 3F-·8H2O⊂2CP2 (right).
To know more details about this fascinating work, were everything have been done with the biggest dedication, please check here!
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in