Recreating a key event of Saccharomyces yeasts evolution in the lab!
Published in Ecology & Evolution

However, it also impairs the ability of hybrids to form new species because hybrids tend merge back with their parental species when backcrossing. Some organisms have access to alternative reproduction mechanisms such as mitotic division. Is there a way for hybrids to recover their fertility without having recourse to sexual reproduction, thus promoting hybrid species formation? We set out to answer this question with an experimental evolution approach. The idea was to generate multiple sets of independent hybrids with different initial levels of fertility. Then we would propagate them asexually for hundreds of generations while minimizing the efficacy of natural selection. This approach would let us observe the broadest spectrum of possible trajectories for the fertility of our hybrids.
We were lucky enough to have access to the perfect study system to undertake this project, the North American yeast Saccharomyces paradoxus lineages. We discovered this wild yeast study system about 7 years ago when I started as a Master student in the lab. It is characterized by having two incipient species that were separated during the last glaciation and have been experimenting secondary contact after glacial retreat. Since then, members of our research group have studied local adaptation, reproductive isolation, natural introgression history, de novo genes emergence and even recurring hybridization with the same yeast system! (Sorry for the shameless plugs)
So here we are, three motivated scientists in training (Mathieu and I were PhD students and Souhir is a Postdoctoral researcher) a supportive PI (thanks Christian). Teamwork often represent a great challenge, but we succeeded in realizing an experiment of an impressive scale. We evolved 672 hybrids lines from four different divergence level between the parental strains. The whole process required:
- 2 years of experimental work, data collection and analyses and manuscript writing.
- more than 8, 000 petris dishes
- about 170 L of culture media
- more than 600 hours of streaking colonies
- more than 200 hours of tetrad dissection
- more than 100 hours of cytometer usage
- hundreds of hours of DNA sequencing and analysis
Overall, we have screened the fertility of more than 200 of the 672 evolution lines at three different timepoints over the course of the evolution. We were happy that all this hard work payed off when we analyzed the data and saw that we had more than one interesting result! First the main result that spontaneous whole-genome duplication restores fertility in interspecific hybrids.
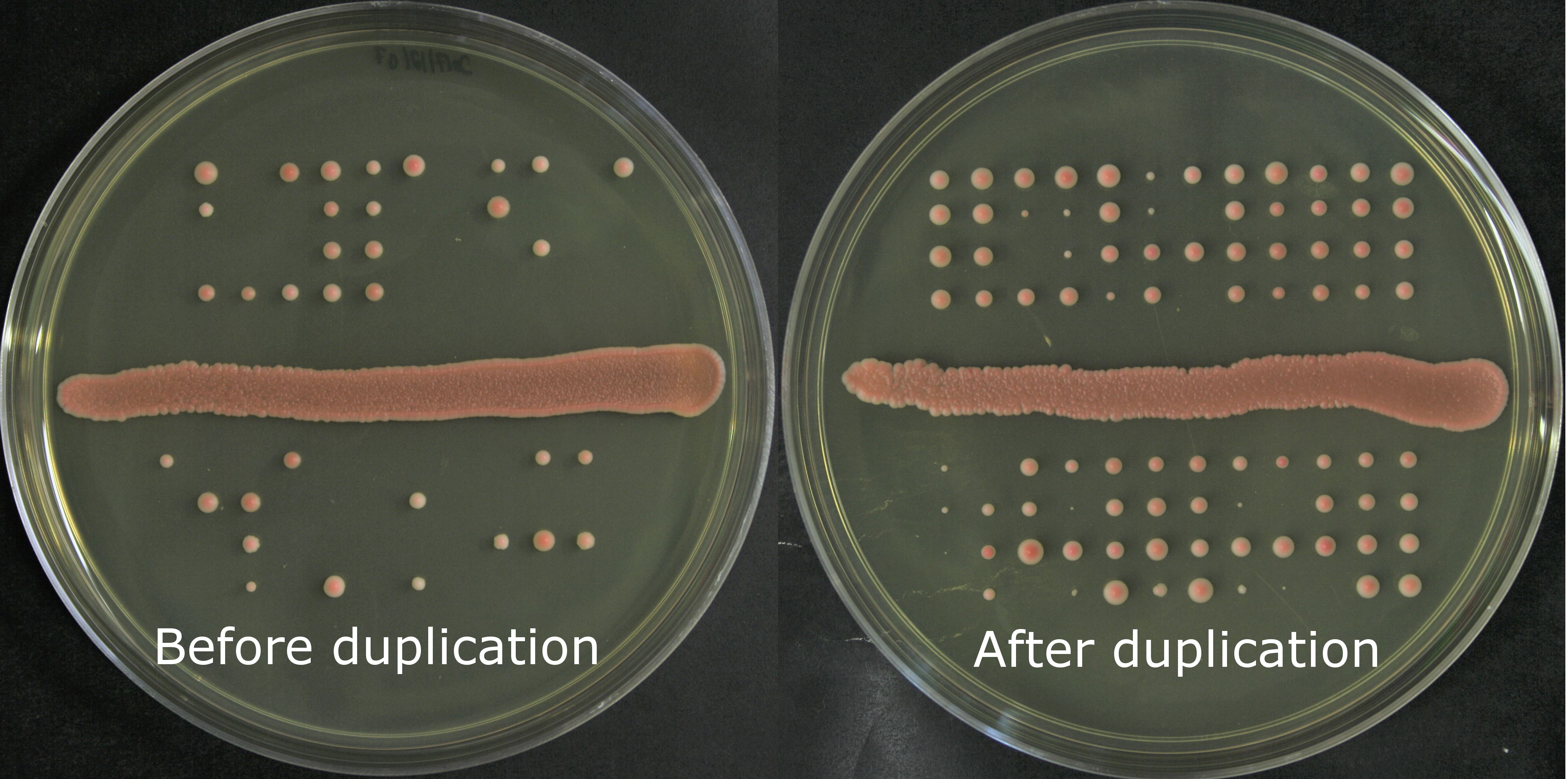
Figure 1: Fertility in yeasts is measured as the proportion of viable spores (haploid yeasts produced by meiosis). Our results show a significant increase in viable spores after the duplication events.
This is of particular interest because hybridization between more or less diverged lineages followed by genome duplication is currently the suggested mechanism that eventually led to the formation of the Saccharomyces genus. So we recreated in the lab something that happened millions of years ago! We also observed that some of the hybrids lost their ability to perform meiosis very early after the initial hybridization event even though they were very good at it right after hybridization.
Another thing we learned is that ploidy instability is both a boon and a curse. Yes the genome doubling helped the originally poorly fertile hybrids to recover to full fertility. However, genome duplication in the initial haploids used in the crosses led to the formation of triploids which had even more reduced fertility than diploid hybrids. However, this ploidy instability seem to have come from particular strains and we still do not know it’s implication in the stability of diploid genomes.
This project may be over, but we feel that we have raised more questions than answers. Duplication may have allowed the recovery of hybrids fertility, but is there recombination between the two divergent genomes during meiosis? Preliminary work performed on the hybrid with the highest divergence suggest that there is none because the two identical chromosomes preferentially pair up with each other at meiosis. This can be a problem because it means that, upon meiosis, the tetraploid generates heterozygous diploid that are unable to mate (yeast cannot mate when heterozygous at the mating type locus) thus unable to regenerate a tetraploid. The nice thing about experimental evolution experiment is that you keep frozen stocks of your lines. This way, you can go back in time or go forward with the strains and basically pursue any questions you still might have! But until the next developments of this story, we are happy to have contributed an original piece of research to the community.
P.S.: I want to thank Souhir Marsit and Mathieu Hénault for their contribution to this blog post. Photo credit for the tetrads in the banner: Mathieu Hénault.
P.P.S.: If you have any insights about the whole ade2 mutants situation described in our paper, please share them with us. We are really curious about what is going on there!
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in