Ring splitting prevents the loading of defective DNA replication machinery
Published in Chemistry and Cell & Molecular Biology
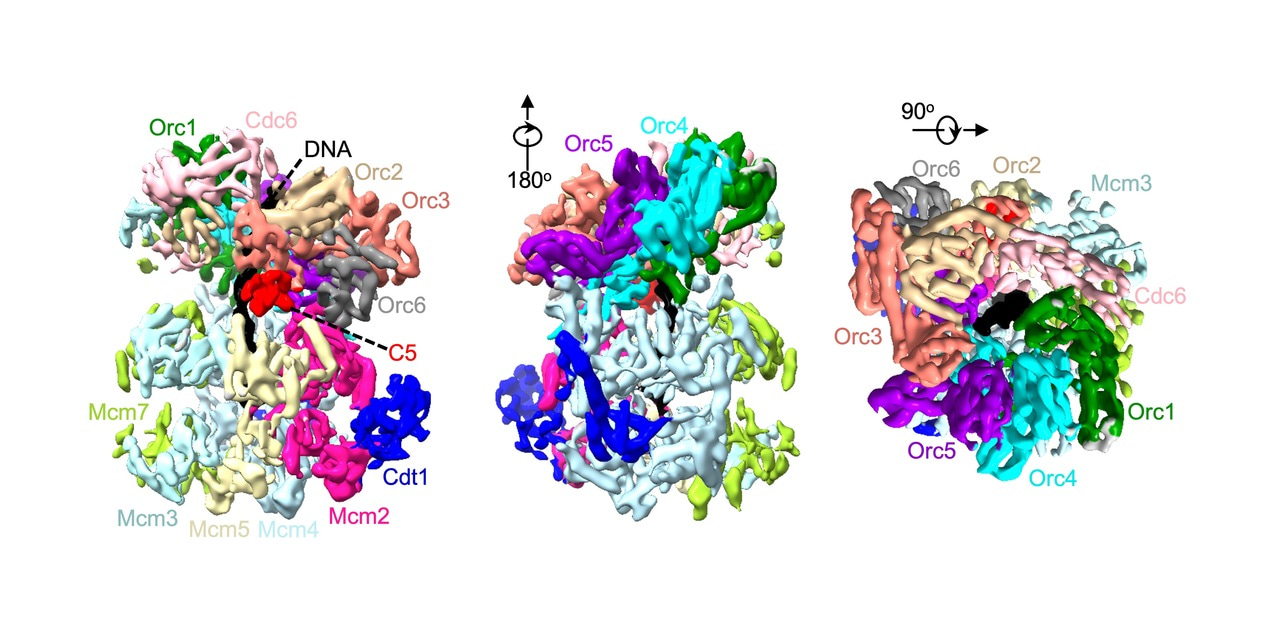
To replicate DNA, you first need to unwind it using a helicase. In eukaryotes the core helicase, MCM2-7, is loaded onto DNA in a multi-step process that requires ATP for energy. We, along with the Li group (Van Andel Institute), have previously published a structure of one of these loading intermediates, the OCCM – named after its protein components: ORC/Cdc6/Cdt1/MCM2-71. In this structure we were unable to see the C-terminus of the Mcm5 subunit (C5). Mcm5, along with Mcm2, creates a gate through which DNA enters the helicase during loading. We also noticed that densities at the Mcm2/Mcm5 gate were “fuzzier” than the rest of the structure. Taking advantage of advances in cryo-EM processing technology, we reprocessed out dataset and found that in 10% of these particles we could finally visual C5. One of the key differences between the two structures was that C5 appeared to be a sticking plaster – stabilising the connection between MCM2-7 and its loader ORC, as well as closing the gap between Mcm2 and Mcm5.
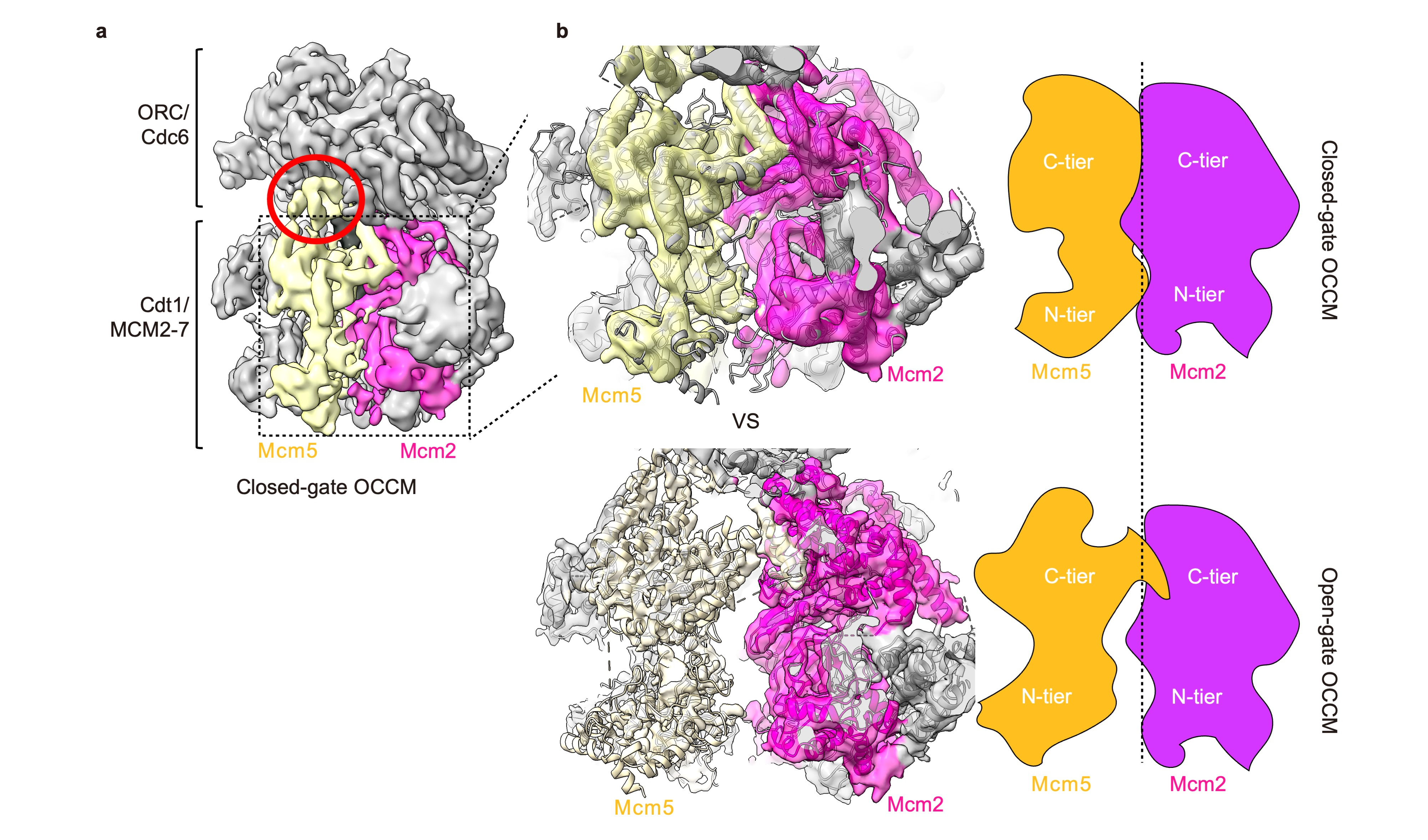
Fig. 1: a Cryo-EM map of the closed-gate OCCM. b The closed-gate OCCM forms a tight interface between Mcm5 (yellow) and Mcm2 (magenta), whereas the open-gate OCCM has a 10-15 Å gap between the subunits.
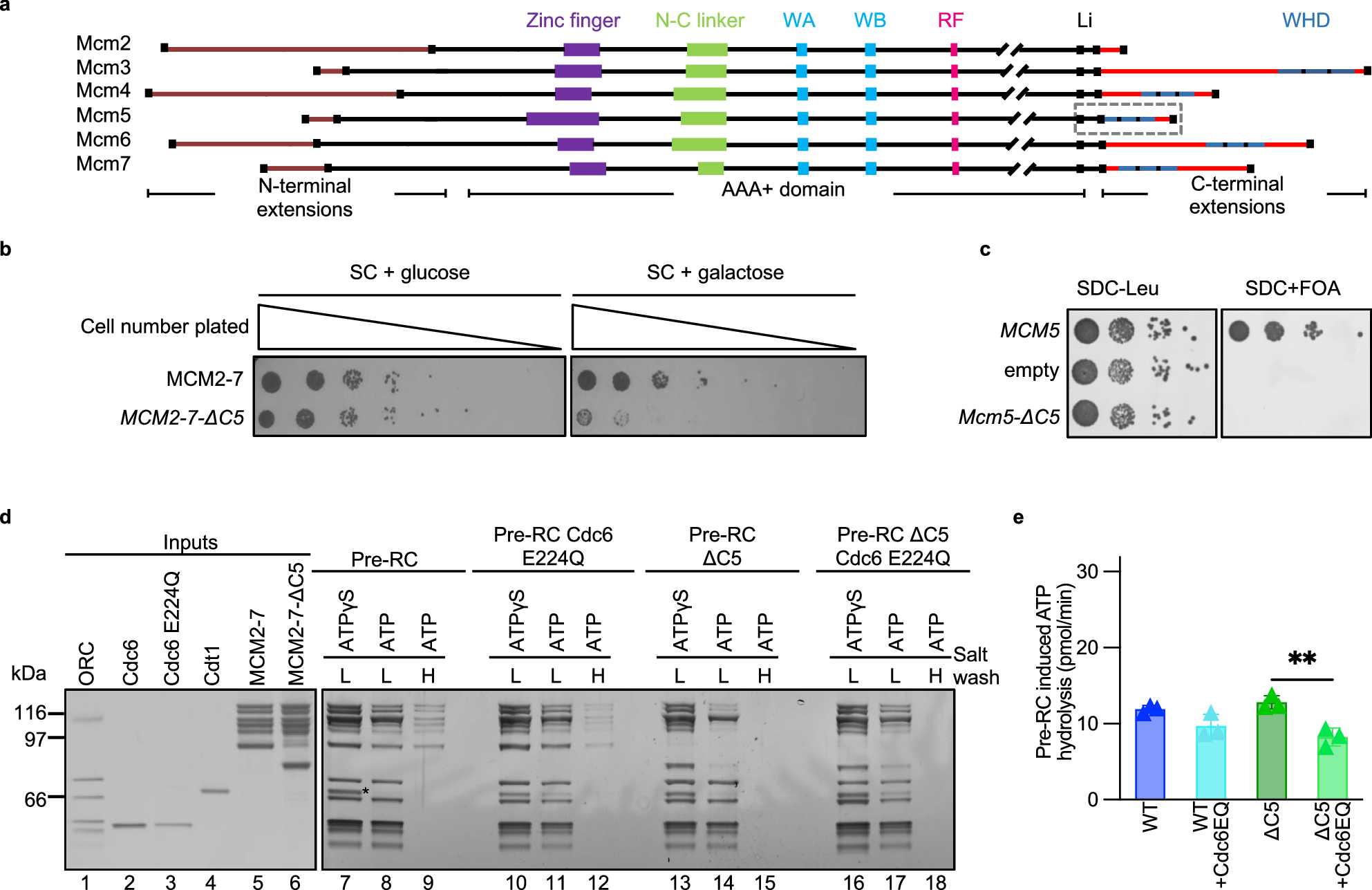
To elucidate the role of C5 in helicase loading, Marta Barbon, a PhD student at the time, truncated the C-terminus of Mcm5 and also produced point mutations. Removing C5 had a big impact on the stability of helicase loading intermediates, suggesting that it could indeed act as a sticking plaster. Mutating C5 induced high levels of ATP hydrolysis. However, this hydrolysis was not being channelled through Cdc6, which was previously identified as a key protein for helicase quality control2-4. A technician in the lab, Audrey Mossler, set-about producing a number of mutant MCM2-7 complexes and testing them in ATPase assays. We determined that the Mcm4/Mcm7 interface was predominantly responsible for the elevated ATP hydrolysis. This was exciting as this interface had previously been implicated in ring-splitting5. I developed a pull-down assay in order to assess ring stability and found that WT MCM2-7 could indeed “spilt” at the Mcm4/Mcm7 interface. This splitting could be prevented using a Mcm4-WB mutant, indicating that ATP hydrolysis at the Mcm4/Mcm7 interface was responsible for the disassembly. To further probe the mechanism, we used our Mcm5 mutants along with phosphorylated ORC - another previously-characterised quality control mechanism that removes Cdt1-MCM2-7, to ensure that helicase loading is blocked in S-phase6. We found that any mutant that caused double-hexamer formation, and therefore ring closure, to fail resulted in splitting of the MCM2-7 hexamer. We were consequently able to connect the role of our C5 sticking plaster with Mcm4 ATP hydrolysis – if C5 can attach to ORC, then Cdt1 is released and helicase loading can proceed, locking the ring onto DNA. If C5 cannot latch, then the ring is destabilised and ATP hydrolysis causes it to split, protecting the cell from loading defective helicases. 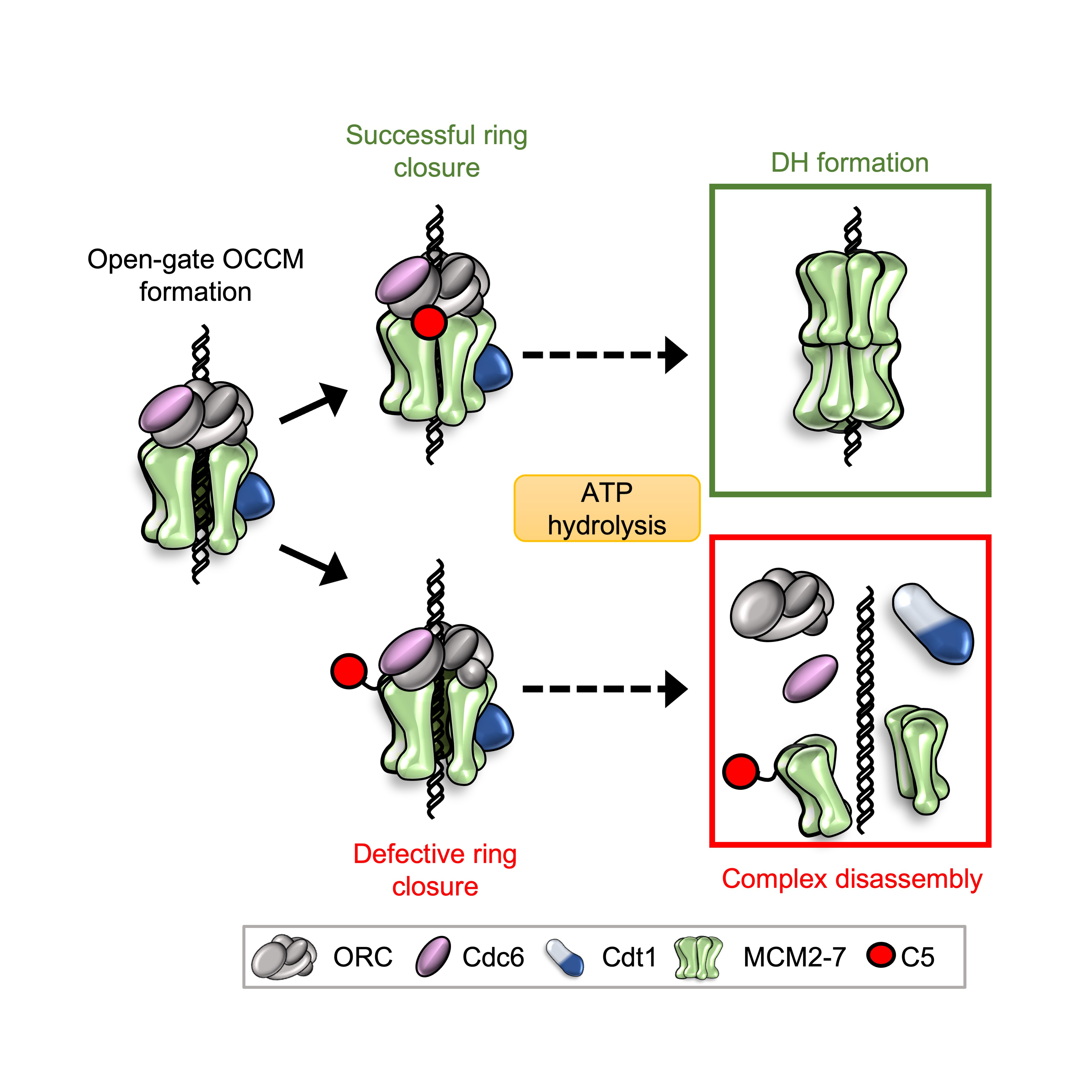
Fig. 2: OCCM formation requires the attachment of C5 (red) for successful ring closure and progression to a an MCM2-7 double hexamer (DH). If C5 cannot attach due to mutations or phosphorylation, then ATP hydrolysis through Cdc6 (pink) and Mcm4 leads to complex disassembly through spitting of the MCM2-7 ring.
1 Yuan, Z. et al. Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol 24, 316-324 (2017). https://doi.org/10.1038/nsmb.3372
2 Coster, G., Frigola, J., Beuron, F., Morris, E. P. & Diffley, J. F. Origin licensing requires ATP binding and hydrolysis by the MCM replicative helicase. Mol Cell 55, 666-677 (2014). https://doi.org/10.1016/j.molcel.2014.06.034
3 Frigola, J., Remus, D., Mehanna, A. & Diffley, J. F. ATPase-dependent quality control of DNA replication origin licensing. Nature 495, 339-343 (2013). https://doi.org/10.1038/nature11920
4 Fernandez-Cid, A. et al. An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly. Mol Cell 50, 577-588 (2013). https://doi.org/10.1016/j.molcel.2013.03.026
5 Guerrero-Puigdevall, M., Fernandez-Fuentes, N. & Frigola, J. Stabilisation of half MCM ring by Cdt1 during DNA insertion. Nat Commun 12, 1746 (2021). https://doi.org/10.1038/s41467-021-21932-8
6 Amasino, A. L., Gupta, S., Friedman, L. J., Gelles, J. & Bell, S. P. Regulation of replication origin licensing by ORC phosphorylation reveals a two-step mechanism for Mcm2-7 ring closing. Proc Natl Acad Sci U S A 120, e2221484120 (2023). https://doi.org/10.1073/pnas.2221484120
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in