RNA-Puzzles Round V: evaluation of RNA structure prediction on 23 targets
Published in Chemistry and Protocols & Methods
The mission
RNA structure prediction remains a major challenge in bioinformatics due to the complexity of RNA folding and the variety of non-covalent interactions involved. The RNA-Puzzles project was launched twelve years ago to provide a community-wide blind prediction experiment to assess the progress of methods for predicting the 3D structure of RNA. RNA-Puzzles provides experimentally determined, unpublished RNA sequences of novel structures for 3D structure prediction. Once the structures are published and available, the RNA-Puzzles team evaluates the accuracy and limitations of different prediction techniques. The results are then published in a joint effort between the structural biology and modelling groups. The aim of the whole process is to stimulate methodological advances in the development of 3D RNA prediction. This briefing summarises the results of RNA Puzzles Round V, highlighting the key findings and challenges faced by the participating teams.
The Discovery
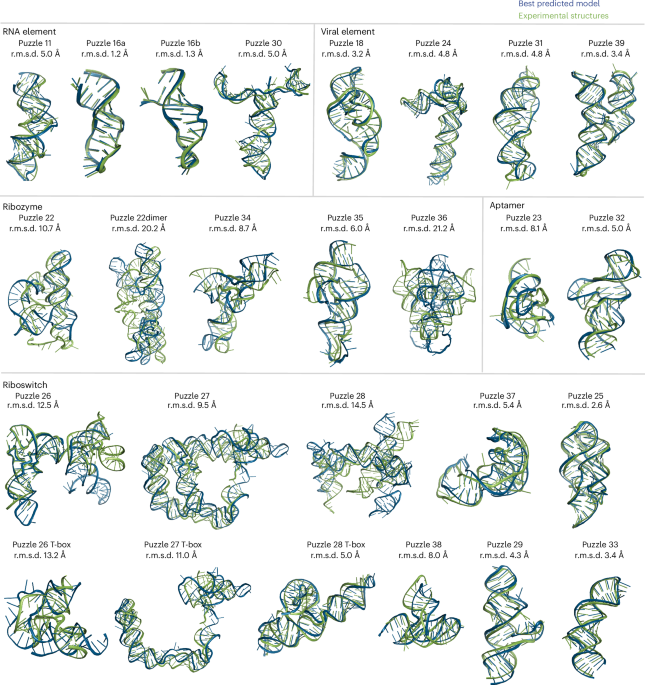
Eighteen groups participated in RNA-Puzzles Round V, predicting the structures of 23 RNA molecules from different functional classes (RNA elements, aptamers, viral elements, ribozymes and riboswitches). The predictions were evaluated using a variety of metrics, including the usual root mean square deviation (RMSD) and precise molecular parameters characteristic of RNA architectures. The metrics include the identification of helix-forming base pairs, recurrent modules formed by non-Watson-Crick pairs, and base-base stacking within and between helices. The analysis revealed key challenges in accurately modelling specific RNA features, such as coaxial stacking and interstrand entanglement. Three of the top four modelling groups in this round also ranked highly in the 2022 CASP15 protein structure prediction competition, suggesting some transferability of modelling expertise. Automatic predictions from web servers, although often less accurate than human-assisted modelling, also contributed useful results and were evaluated separately. Several specific RNA-puzzles proved exceptionally difficult to model, highlighting the challenges of predicting complex RNA architectures and ligand interactions.
The Implication
RNA-Puzzles Round V demonstrates continued progress in RNA 3D structure prediction, but also highlights significant challenges that remain. The identification of areas for improvement (such as modelling non-Watson-Crick interactions and predicting complex RNA folds) directs future research efforts towards improved imaginative algorithm development and computational resources. The continued participation of top groups from CASP suggests the potential for cross-disciplinary advances in structure prediction methodologies. Although artificial intelligence tools are powerful for abstracting and generalising folding rules extracted from large datasets, the available data for RNA structures is still limited. A combination between AI and a good understanding of RNA folding rules by human experts could be part of future directions in this field.
The double-blind nature of the competition ensures an objective evaluation and facilitates a fair comparison of prediction methods. The release of data (experimental structures) and results (predictions) will be open access, furthering the goals of data sharing and community-driven progress in the field.
Future rounds of RNA-Puzzles will benefit from the identification of improved methods for assessing model quality and the refinement of protocols to address the challenges encountered in Round V. The incorporation of additional experimental data, such as chemical probing data, will also help to generate better constraints for prediction methods.
Behind the paper
I started my research career in RNA structure modelling when I joined Eric Westhof’s laboratory in Strasbourg (France). He had already started organising RNA-Puzzles and I agreed to continue the project. RNA-Puzzles is a very exciting project for me because it aims to understand the capabilities and bottlenecks of 3D RNA structure modelling. Over the past decade, we have published four rounds of summary reports highlighting the limitations of prediction (e.g. non-Watson-Crick base pairs and structural modules). Together with our colleagues in the community, we have also organised several RNA Puzzles meetings to better share and advance the field (2016 in Strasbourg, 2018 in Warsaw, 2023 in Montreal). During the COVID pandemic, we realised that our prediction could help with a potential therapy and modelled the untranslated RNA regions of SARS-CoV-2 with community efforts. This year, the Nobel Prize was awarded to the developers of AlphaFold, the best AI tool for protein structure prediction. However, the problem of predicting the 3D structure of RNA is not yet solved. Further advances in this field could benefit RNA therapies, including mRNA vaccines, RNA aptamers and RNA-targeted small molecule drugs. Z.M.
Follow the Topic
-
Nature Methods

This journal is a forum for the publication of novel methods and significant improvements to tried-and-tested basic research techniques in the life sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Methods development in Cryo-ET and in situ structural determination
Publishing Model: Hybrid
Deadline: Jul 28, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Was RNA created before the formation of DNA?