Secrets in the Light: Unveiling eccDNA in the Rice Genome and Its Implications
Published in Plant Science
The genome, an intricate and fluid network, perpetually adapts to a myriad of external and internal stimuli, standing at the core of evolutionary innovation and the vast tapestry of biodiversity. Within this genomic puzzle lies one of its most enigmatic pieces: extrachromosomal circular DNA (eccDNA). Existing independently from the primary chromosomal framework, eccDNA was initially discovered in the cells of eukaryotic organisms. This unique form of DNA spans a broad spectrum of functions, from facilitating gene amplification to playing a crucial role in the regulation of gene expression, thereby contributing significantly to the pool of genetic diversity.
Despite its identification in a variety of human tissues and certain dicotyledonous plants, the exploration of eccDNA, especially in the realm of plant biology, remains in its nascent stages. This gap in knowledge beckons a deeper investigation into its origins, mechanisms, and implications.
Leveraging advanced methodologies such as Circle-seq, PCR, and third-generation sequencing technologies, our research team has shed light on the universal characteristics of eccDNA formation, reinforcing the hypothesis of its widespread occurrence across different species. Specifically, our studies have unveiled two distinct pathways through which eccDNA forms in rice, a staple food crop. One pathway, characterized by the presence of Direct Repeats, suggests a possible link to the alternative end-joining (Alt-EJ) repair mechanism. Moreover, our findings propose that the process of eccDNA formation can result in chromosomal deletions, offering a novel lens through which to view inheritable genomic alterations.
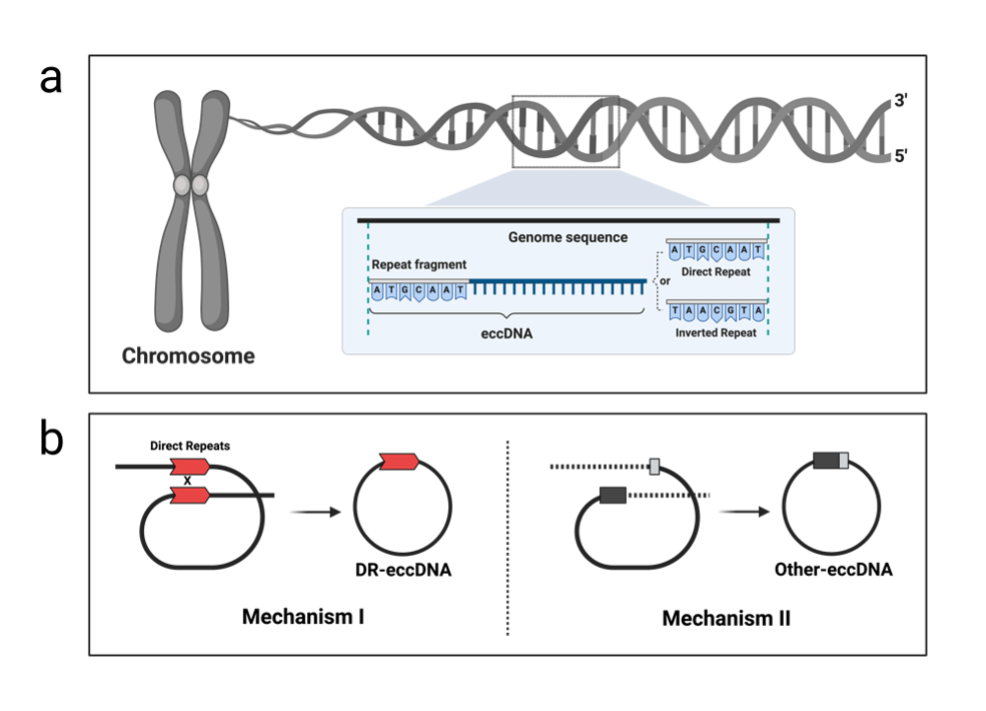
Schematic diagram of the eccDNA formation mechanism induced by Direct Repeats
Importantly, we delineate that eccDNA influences gene expression indirectly, adding a layer of complexity to our understanding of genomic regulation. A striking discovery within our research is the pronounced accumulation of eccDNA in the leaf tissues of rice plants, potentially a defense response to DNA damage caused by intense light exposure and radiation stress. This suggests that eccDNA could serve as a crucial biomarker for assessing DNA damage in rice, presenting an invaluable tool for breeding programs aimed at enhancing stress tolerance in crops.
Such advancements are imperative as we face the growing challenge of climate change, with an urgent need for crops that can thrive under adverse environmental conditions. The insights gleaned from our exploration into the rice genome and the role of eccDNA in genomic adaptation underscore the potential of eccDNA to influence crop resilience, yield, and the overall improvement of agricultural practices.
In sum, our journey into the largely unexplored domain of eccDNA within the rice genome uncovers its pivotal role in driving genomic evolution and adaptation. These pioneering insights pave the way for future research endeavors in rice breeding, promising not only to enrich our understanding of plant genomics but also to revolutionize agricultural strategies in the face of global climatic shifts. Through this lens, we invite the scientific community to further unravel the mysteries of eccDNA, fostering innovations that will sustain and enhance crop productivity for future generations.
For details, please refer to our paper at https://doi.org/10.1038/s41467-024-46691-0.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in