Sensing relatedness to fine-tune antibiotic production
Published in Microbiology

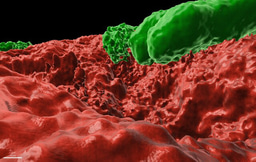
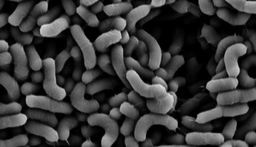
Bacillus subtilis and its related species, represent an intriguing model of biocontrol agents and PGPR, which is of great relevance to ecology and agriculture. B. subtilis, B. amyloliquefaciens, B. velezensis and B. mojavensis, are widely used for the biological control of bacterial, viral and fungal plant pathogens and as plant growth promoting agents1. Similarly to probiotics, these beneficial microbiome members were suggested to promote the growth and wellness of their plant host, and thereby can reduce the use of hazardous pesticides in agriculture. About 5% of the genome of these beneficial bacteria is dedicated to the synthesis of antimicrobial molecules by non-ribosomal peptide synthetases (NRPS) or polyketide synthases (PKS/NRPS)2.
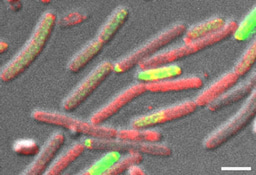
In this work, we first assessed the contribution of four such antibiotics to the competition between Bacillus species sharing the same niche with Bacillus subtilis comparing the single, double and quadruple mutants in competition with their parental strain. Our results clearly demonstrated that the studied NPR and PKS biosynthetic clusters, which are responsible for the biosynthesis of surfactin, bacillaene, bacilysin and plipastatin are not redundant. Instead, each NRP/polyketide contributed to the capacity of Bacillus subtilis to successfully eliminate its competitors. Furthermore, B. subtilis was inert to its own clade members. Neither of the measured antibiotics was toxic to. B. subtilis, and B. amyloliquefaciens strains. Using a mathematical model we could decipher the level of synergy between bacillaene (a selective inhibitor for protein synthesis)3-7 and surfactin (a powerful surfactant associated with membrane leakage)8-11 ,and to determine that although bacillaene and surfactin contribute most to the elimination of non-self-competitors, all four antibiotics are synergistic.
We followed up to ask whether bacteria will express antibiotics while in proximity to resistant competitors. From ecological point of view, this type of wasteful antibiotic production is expected to be deleterious to the bacteria in a competitive environment, reducing their capacity to grow due to high investment in antibiotic production, while potentially selecting for antibiotic resistance genes in the immediate environment. Quite elegantly, all antibiotics are induced only when bacteria are in proximity to a sensitive competitor. The induction of antibiotic production by sensitive competitors requires the transcriptional regulator ComA- a master regulator that also mediated genetic competence12. This indicates an unexpected link between horizontal gene transfer and antibiotic production- when bacterial communities (both antibiotic production and sensitivity sensing require a critical mass of bacteria) are in proximity to phylogenetically unrelated and thereby sensitive competitor, at least within the Bacillus phylum, they will activate antibiotic production. On the same time they will also transcribe the set of genes required for uptake of the foreign DNA from the lysed bacterium.
To learn more on sensitivity sensing and the ecological tradeoff between antibiotic production and growth, please visit our paper: https://www.nature.com/articles/s41467-021-27904-2
1 Hou, Q. & Kolodkin-Gal, I. Harvesting the complex pathways of antibiotic production and resistance of soil bacilli for optimizing plant microbiome. FEMS Microbiol Ecol 96, doi:10.1093/femsec/fiaa142 (2020).
2 Caulier, S. et al. Overview of the Antimicrobial Compounds Produced by Members of the Bacillus subtilis Group. Front Microbiol 10, 302, doi:10.3389/fmicb.2019.00302 (2019).
3 Erega, A. et al. Bacillaene Mediates the Inhibitory Effect of Bacillus subtilis on Campylobacter jejuni Biofilms. Appl Environ Microbiol 87, e0295520, doi:10.1128/AEM.02955-20 (2021).
4 Muller, S. et al. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl Environ Microbiol 80, 5603-5610, doi:10.1128/AEM.01621-14 (2014).
5 Calderone, C. T., Bumpus, S. B., Kelleher, N. L., Walsh, C. T. & Magarvey, N. A. A ketoreductase domain in the PksJ protein of the bacillaene assembly line carries out both alpha- and beta-ketone reduction during chain growth. Proc Natl Acad Sci U S A 105, 12809-12814, doi:10.1073/pnas.0806305105 (2008).
6 Reddick, J. J., Antolak, S. A. & Raner, G. M. PksS from Bacillus subtilis is a cytochrome P450 involved in bacillaene metabolism. Biochem Biophys Res Commun 358, 363-367, doi:10.1016/j.bbrc.2007.04.151 (2007).
7 Butcher, R. A. et al. The identification of bacillaene, the product of the PksX megacomplex in Bacillus subtilis. Proc Natl Acad Sci U S A 104, 1506-1509, doi:10.1073/pnas.0610503104 (2007).
8 Lopez, D., Fischbach, M. A., Chu, F., Losick, R. & Kolter, R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A 106, 280-285, doi:10.1073/pnas.0810940106 (2009).
9 Straight, P. D., Willey, J. M. & Kolter, R. Interactions between Streptomyces coelicolor and Bacillus subtilis: Role of surfactants in raising aerial structures. J Bacteriol 188, 4918-4925, doi:10.1128/JB.00162-06 (2006).
10 Fenibo, E. O., Ijoma, G. N., Selvarajan, R. & Chikere, C. B. Microbial Surfactants: The Next Generation Multifunctional Biomolecules for Applications in the Petroleum Industry and Its Associated Environmental Remediation. Microorganisms 7, doi:10.3390/microorganisms7110581 (2019).
11 Rosenberg, G. et al. Not so simple, not so subtle: the interspecies competition between Bacillus simplex and Bacillus subtilis and its impact on the evolution of biofilms. NPJ Biofilms Microbiomes 2, 15027, doi:10.1038/npjbiofilms.2015.27 (2016).
12 Dubnau, D., Hahn, J., Roggiani, M., Piazza, F. & Weinrauch, Y. Two-component regulators and genetic competence in Bacillus subtilis. Res Microbiol 145, 403-411, doi:10.1016/0923-2508(94)90088-4 (1994).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in