Sharing is caring
Published in Microbiology

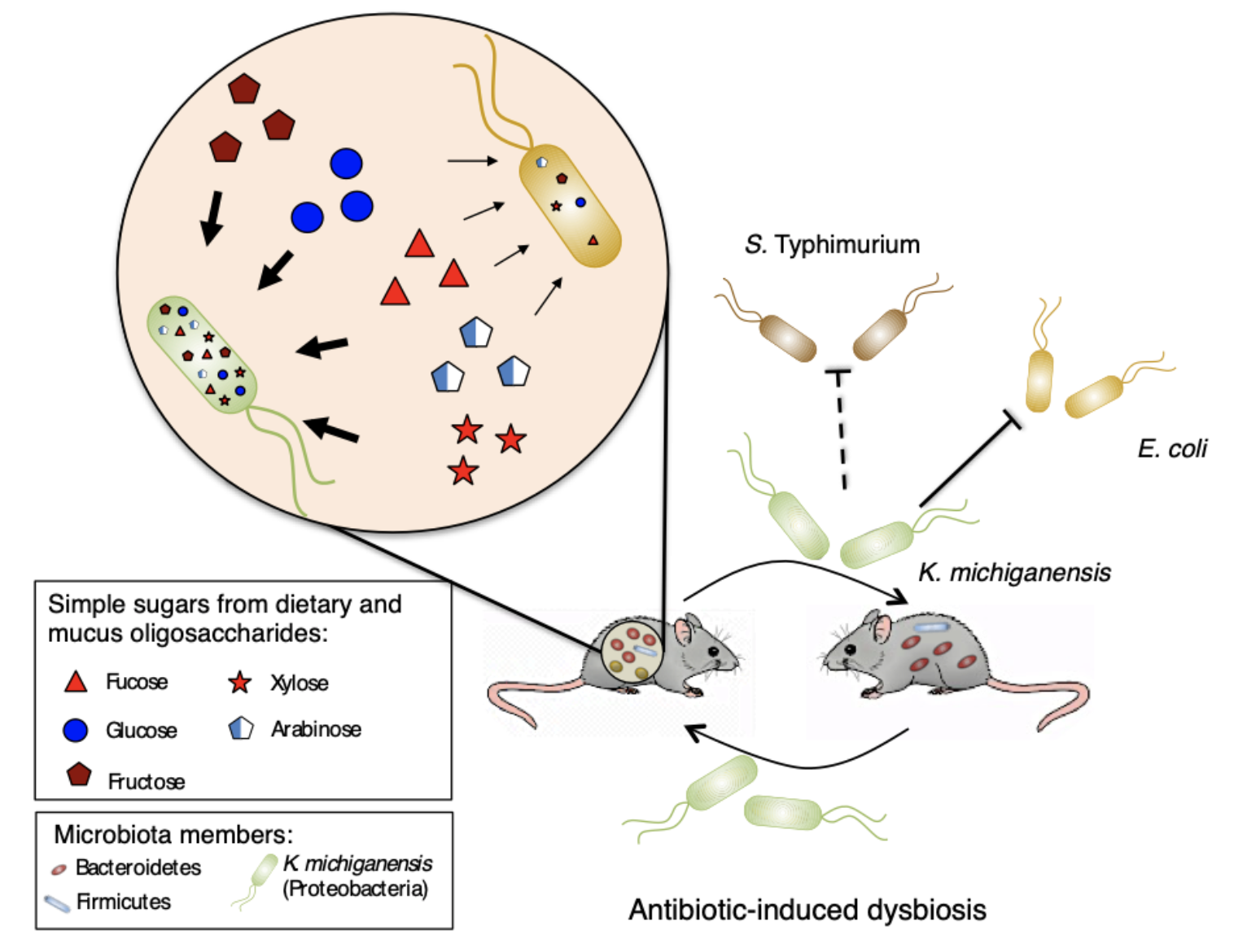
In our latest work, now published in Nature Microbiology, we studied the effect of sharing microbiota among hosts in gut recolonization post-antibiotic-induced dysbiosis and identified a keystone microbiota member that is sufficient to restore colonization resistance against Enterobacteriaceae by nutrition competition.
Antibiotics have revolutionized medical practices dealing with bacterial infections. However, there are compiling evidences of the tremendous impact of antibiotics in our resident gut microbiota by massively reducing its diversity. Even after stopping antibiotic treatment, recovery of microbiota diversity is a slow process with extinction of key commensal bacterial species, taking several weeks or months. A reduction in microbiota diversity opens the door for opportunistic pathogenic bacteria that, with reduced competition, can colonize the gut and compromise host well-being. Increasing knowledge on this recolonization process is important to promote host health.
In our group, while studying bacterial signaling effect on mouse gut microbiota recovery after streptomycin exposure, we stumbled upon a crucial issue: no recovery whatsoever… In 2016, we met Kerwyn Casey Huang (Stanford University) at a conference, who was also studying factors affecting microbiota recovery from antibiotics. While discussing our work, the hypothesis arose that microbiota sharing among hosts could promote recovery from treatment. This was the beginning of a collaboration between our lab in Portugal and Huang lab in the USA. We studied in parallel the effect of co-housing in recovery from dysbiosis, using two different antibiotics. While we studied the effect of streptomycin, Huang lab focused on the effect of ciprofloxacin. Both experiments showed a beneficial effect of co-housing with increased diversity and microbiota similarity. However, unlike in the ciprofloxacin study, in the streptomycin experiment microbiota diversity never reached the levels observed before treatment. This prompted us to test if microbiota functions had been recovered by co-housing after streptomycin treatment. Indeed, while testing colonization resistance to an invading Escherichia coli, as a microbiota function commonly impaired by antibiotic treatment, we observed it was increased in co-housed mice. Using a classical microbiology technique to assert E. coli loads by plating and counting colonies, we noticed unexpected morphologically different colonies. These colonies were identified as Klebsiella michiganensis and were only present in mice with increased colonization resistance to E. coli. Later, in vivo experiments confirmed that K. michiganensis was sufficient to provide colonization resistance and displacement of E. coli.
Since K. michiganensis was undetectable both before and during antibiotic treatment, alarms were raised. Did we contaminate our mice after streptomycin treatment? Or is K. michiganensis a low abundant member of the untreated microbiota? To unravel this dilemma, we tested 28 cohorts of untreated mice from our rodent facility for the presence of Klebsiella, developing an enrichment-based method. This revealed that Klebsiella was a low abundant non-prevalent member of our mice gut microbiota, appearing in 25% of cohorts. Because we never detected resistance to streptomycin in Klebsiella, we suspected that the sharing of microbiota in co-housed mice was responsible for maintaining Klebsiella in the gut when present before treatment. To test this hypothesis, we used one cohort that scored positive and another cohort that scored negative for Klebsiella presence with our enrichment method. As expected, after streptomycin treatment, Klebsiella was only present in co-housed mice of the cohort that had scored positive for Klebsiella before treatment. This shows that microbiota sharing can be important for maintaining this low abundant beneficial member of our gut.
Intrigued by this uncharacterized interaction between K. michiganensis and E. coli, we performed a battery of studies to uncover the underlying mechanism. After discarding mechanisms based on contact-dependent killing and inhibitory compounds, we reasoned that the higher nutrient consumption capacity of K. michiganensis was the most suitable strategy to outcompete E. coli. However, this fundamental cell biology process turned out to be harder than expected to test in vivo. The difficulty lies in the impossibility of reducing the gut environment to a single nutrient that we could impair K. michiganensis to consume. Therefore, the redundant and higher nutrient consumption capacity of K. michiganensis would demand several parallel mutations to impair it. As we could not test this in vivo competition on the winner, we decided to test it on the loser, E. coli, by conferring it an advantage in a form of an exclusive nutrient, inaccessible to K. michiganensis. This strategy successfully abolished K. michiganensis advantage over E. coli, enabling both species to colonize the gut at high loads, confirming competition for nutrients as the underlying mechanism of this inhibition.
We hope that our work can lead to new efforts on the discovery of important commensal gut bacteria against invading pathogens.
Check out our paper here: https://www.nature.com/articles/s41564-019-0658-4
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
I am super proud of this beautiful study from my student Rita Oliveira. It is a beautiful story all the way from studying the effect of antibiotics on the microbiota, to the identification of a keystone microbiota member with a major role in protection against Enterobacteriaceae, to the identification of the mechanism involved in this protection.
What a lovely comment !! Well done on the research and paper, and thanks for sharing the story on our community :)
Thank you Ruth