Small genomes, big appetites: amino acid auxotrophy in the bacterial world
Published in Ecology & Evolution

There are 9 essential amino acids that humans cannot produce, and thus we rely on the food we eat to provide us with those amino acids. In the microbial world, there is a divide between those that can produce all amino acids, and those that (like us) rely on their diet to obtain them. In certain environments, resources are so scarce or variable that microbes cannot rely on obtaining amino acids directly from their surroundings – they need to produce all amino acids on their own. Other environments are so rich that some microbes can afford not to produce amino acids (so-called amino acid auxotrophs), because these can be readily obtained from their surroundings. Synthesizing amino acids comes at a metabolic cost so if that cost can be avoided, we would expect microbes to lose the ability to synthesize certain amino acids over time. Why pay for something that you can get for free?
We were motivated to study bacterial amino auxotrophies because we know surprisingly little about the prevalence of amino acid auxotrophies across the broad diversity of bacteria, and whether there are certain habitats that select for auxotrophic bacteria remains unclear. We wanted to know how common auxotrophy is across bacteria, which bacteria are auxotrophic, where auxotrophic bacteria are most likely to be found, and what other bacterial traits are associated with amino acid auxotrophy (see our paper - https://www.nature.com/articles/s41467-023-43435-4). Solving these mysteries would provide important insights into the most fundamental aspects of microbial adaptation that make them so successful across Earth’s ecosystems.
The capacities for producing amino acids and other essential compounds are imprinted in the genome of any organism. If we know the genes involved in the production of amino acids, we can infer the capacity to produce these amino acids based on the presence of these genes in any given genome. This is not an easy task, because many of the genes involved in these metabolisms remain unresolved. However, novel genome annotation tools allow us to make inferences about amino acid metabolism in bacteria. In this study, we used information stored in >26,000 genomes to predict the capacity for amino acid production across a broad diversity of bacteria, and analyzed the capacity for amino acid synthesis in bacterial communities from across 12 common microbial habitats. These habitats included soils, oceans, lakes, human and plant microbiomes, water treatment plants, and even foods such as cheese or sourdough (>3,800 samples).
We know that microbes form networks of interactions where they often exchange amino acids. This had led to the expectation that the inability to produce amino acids is widespread in the bacterial world, but is this really true? Probably not. We found that most bacteria (78%) are likely able to produce all the amino acids they need to grow (Figure 1A), although amino acid auxotrophy can be observed across members of most bacterial families. Consistent with evolutionary theory, we found that environments where we expect to see a higher availability of amino acids favor bacteria that cannot synthesize amino acids on their own. For example, fermented foods and the human gut are environments rich in amino acids, and these environments typically harbor many auxotrophic bacteria. The reason is simple – if a microbe has ready access to an ‘all-you-can-eat’ buffet that is open 24/7, where food is plentiful and never runs out, there is no advantage for a microbe to produce the amino acids it can easily get from its surroundings.
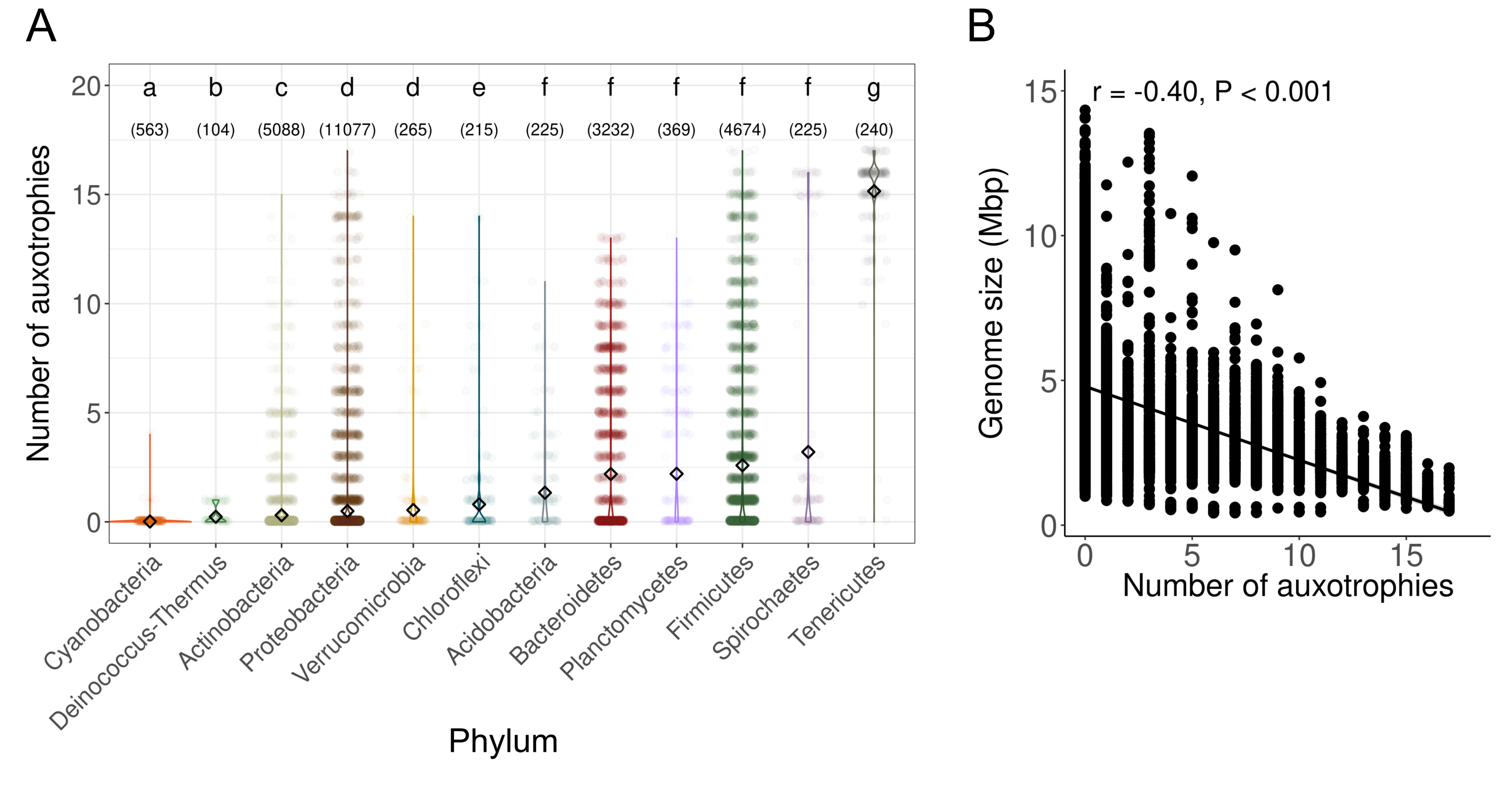
Figure 1. A) Prevalence of amino acid auxotrophy in bacteria. B) Bacteria with a larger number of amino acid auxotrophies tend to have smaller genomes, in agreement with the genome streamlining hypothesis.
Being unable to produce amino acids was not an exclusive feature of those microbes living in fermented food products or our guts. Mycoplasmas that parasitize our cells and cause important diseases are also often auxotrophic as they can obtain amino acids directly from the intracellular environment. We also observed that bacteria which are adapted to predate on other bacteria lacked many genes for amino acid production, including members of the Bdellovibrionaceae. Again, they can obtain these essential compounds from their victims, so they have evolved to avoid the energetic burden of synthesizing amino acids on their own.
Do bacteria that produce all their amino acids have different characteristics from bacteria that rely on the environment to obtain their amino acids? We found numerous characteristics that distinguish these two broad categories of bacteria. Most importantly, bacteria producing all their amino acids had larger genomes than bacteria relying on external amino acid supply (Figure 1B). This indicates that the loss of genes for amino acid production might be just one aspect of a broader adaptive strategy involving the loss of genes associated with other metabolic capabilities. This process is called genome streamlining, a general strategy that allows bacteria to minimize energetic and metabolic costs. Consistent with this view, we found that bacteria that can produce all amino acids are also more competent at lipid, carbohydrate, and other important metabolisms than bacteria which cannot produce all of the necessary amino acids on their own. Our results highlight the importance of a microbe’s surrounding environment in defining its survival strategies, showing that in most environments the best strategy is to be self-sufficient, but that in nutrient-rich and host-associated environments genome streamlining can be a very successful strategy. In the microbial world, bacteria with the smallest genomes have the biggest appetites.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Congrats, Josep!