
Camelids, including alpacas express heavy chain-only antibodies. The lack of the light chain results in smaller size, but they largely retain antigen specificity and affinity comparable to conventional antibodies. Since they are expressed from a single gene, they are easier to clone and characterise and can be further reduced in size to about one tenth that of conventional antibodies. This small size and other characteristics have made these so-called ‘nanobodies’ attractive as molecular laboratory tools, as well as therapeutic and diagnostic agents. Since 2017, Leo Hanke in my research group at the Karolinska Institute in Stockholm has been developing a panel of nanobodies against chikungunya virus proteins and various host factors. Leo established a nanobody identification procedure which was working like a well-oiled engine by January 2020.
At that point it was becoming clear that the novel coronavirus ‘2019-nCoV’, since renamed SARS-CoV-2, was showing no signs of being restricted to a single region. In discussion with our colleague Ben Murrell, we decided to temporarily shelve our CHIKV projects and turn to the development of nanobodies targeting the SARS-CoV-2 spike protein with the aim to block infection. The combination of Ben’s experience with the study of HIV neutralising antibodies and Leo’s skills with alpaca nanobodies gave us confidence that we could efficiently generate SARS-CoV-2 neutralising nanobodies in a relatively short time.
Our partners, PreClinics Research Farm had a handsome alpaca, named Tyson (pictured above), ready for us and we moved quickly to set up an immunisation with S1 protein subunit. We then cloned and expressed a smaller spike fragment containing only the receptor binding domain (RBD) for booster immunisations. A week before the 60-day immunisation was complete, we found that a sample of Tyson’s serum was very clearly positive for RBD binding antibodies. And moreover, a SARS-CoV-2 pseudovirus neutralisation assay revealed that the serum contained just what we were looking for – antibodies that could block infection! But if any of those antibodies were of the single chain variety, we did not yet know.
Leo cloned all nanobody expressing sequences from Tyson’s B cells into a bacteriophage library and started panning for those nanobodies that bound to the RBD. This procedure lead to the enrichment of several nanobodies, but one in particular stood out. A strong binder, clearly enriched after phage display, expressing well in bacteria and displaying good solubility and heat stability. We called it Ty1 and continued with downstream analyses with it alone. Next generation sequencing of the bacteriophage libraries revealed that Ty1 had been enriched 10,000-fold by just two rounds of phage display. The chosen one! And it certainly lived up to its promise.
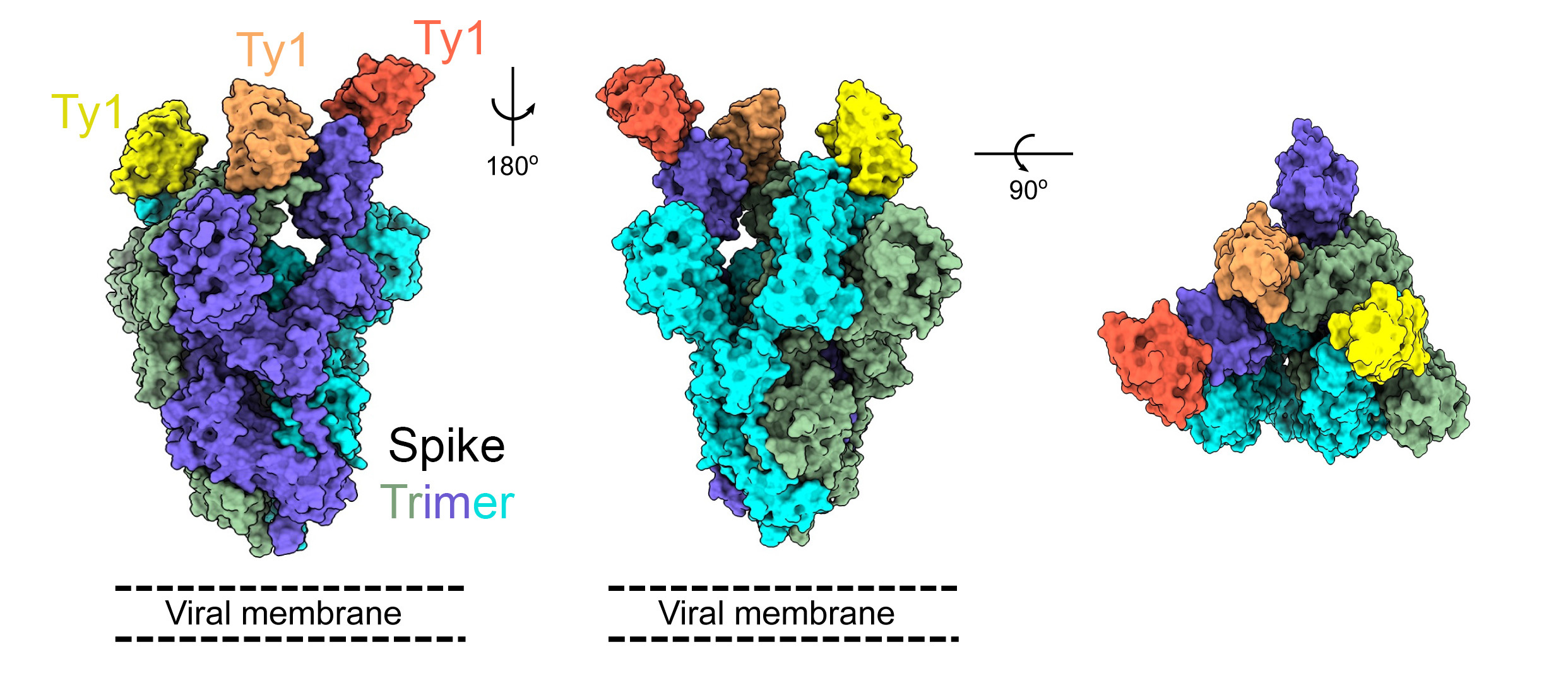
Ty1 neutralises infection with SARS-CoV-2 and, when expressed as a bivalent molecule using an Fc domain, is many times more potent. Our colleague Martin Hällberg solved the structure of the bound complex by cryo-electron microscopy, providing a structural understanding of the potent neutralisation. We propose that Ty1 could be used as a therapeutic agent against SARS-CoV-2 infection.
It has also proven useful as a lab tool. Directly conjugated to a fluorophore, Ty1 lends itself very well to fluorescence microscopy and flow cytometry, giving strong signals in SARS-CoV-2 infected cells with low background. In addition, we have already shared Ty1 protein with at least a dozen laboratories worldwide for use as SARS-CoV-2 neutralisation assay standard. Contact us if you’re interested to use Ty1 in your experiments.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
This is really great work! I hope the success story forTy1 will continue.
Thanks very much Stefanie!