Streamlining DNA Mutation Detection: A Breakthrough with Phosphorothioated DNA-guided Endonuclease Excision
Published in Bioengineering & Biotechnology
Challenges in DNA Mutation Detection:
Molecular diagnostics of cancer has underscored the significance of driver genetic alterations in tumor growth and metastasis1, 2. Although genetic variations hold abundant diagnostic and therapeutic clues for cancer patients, detecting mutations in cfDNA proves to be a major challenge due to their typically low levels. One of the primary obstacles in sensitive detection of low-level altered DNA is the intense background noise arising from an abundance of wild-type (WT) DNA strands.
Currently, DNA mutation detection methods fall into two categories: direct detection methods and enrichment-based methods. While direct detection methods like qPCR3, droplet digital PCR (ddPCR) 4, and sequencing5 have detection limits greater than 0.1%, enrichment techniques encounter hurdles in clinical applications due to complex probe design, temperature sensitivity, cost of auxiliary instruments/reagents, and limited general applicability.
The Precise DNA Excision Tool:
Our major breakthrough involved developing a precise DNA excision tool based on a competitive DNA-binding-and-digestion mechanism. This mechanism is facilitated by deoxyribonuclease I (DNase) guided by single-stranded phosphorothioated DNA (sgDNase), enabling the removal of WT DNA strands (Fig. 1a). By guiding DNase with appropriate sspDNA, we achieved single-nucleotide resolution, surpassing our previously reported DNase/sspDNA complexes6.
Single-nucleotide Resolution:
We first validated the sequence specificity of sgDNase by fluorescent probe method, high performance liquid chromatography analysis and gel electrophoresis analysis respectively. As shown in Figure 1b-d, under the guidance of sspDNA with a sequence fully complementary to the WT DNA sequence, sgDNase could rapidly digest WT DNA strands with high selectivity. In contrast, the majority of the mutant-type DNA strands (MT) were retained. Moreover, the results demonstrate the discrimination effect of sgDNase on different types of mismatches across various genes.
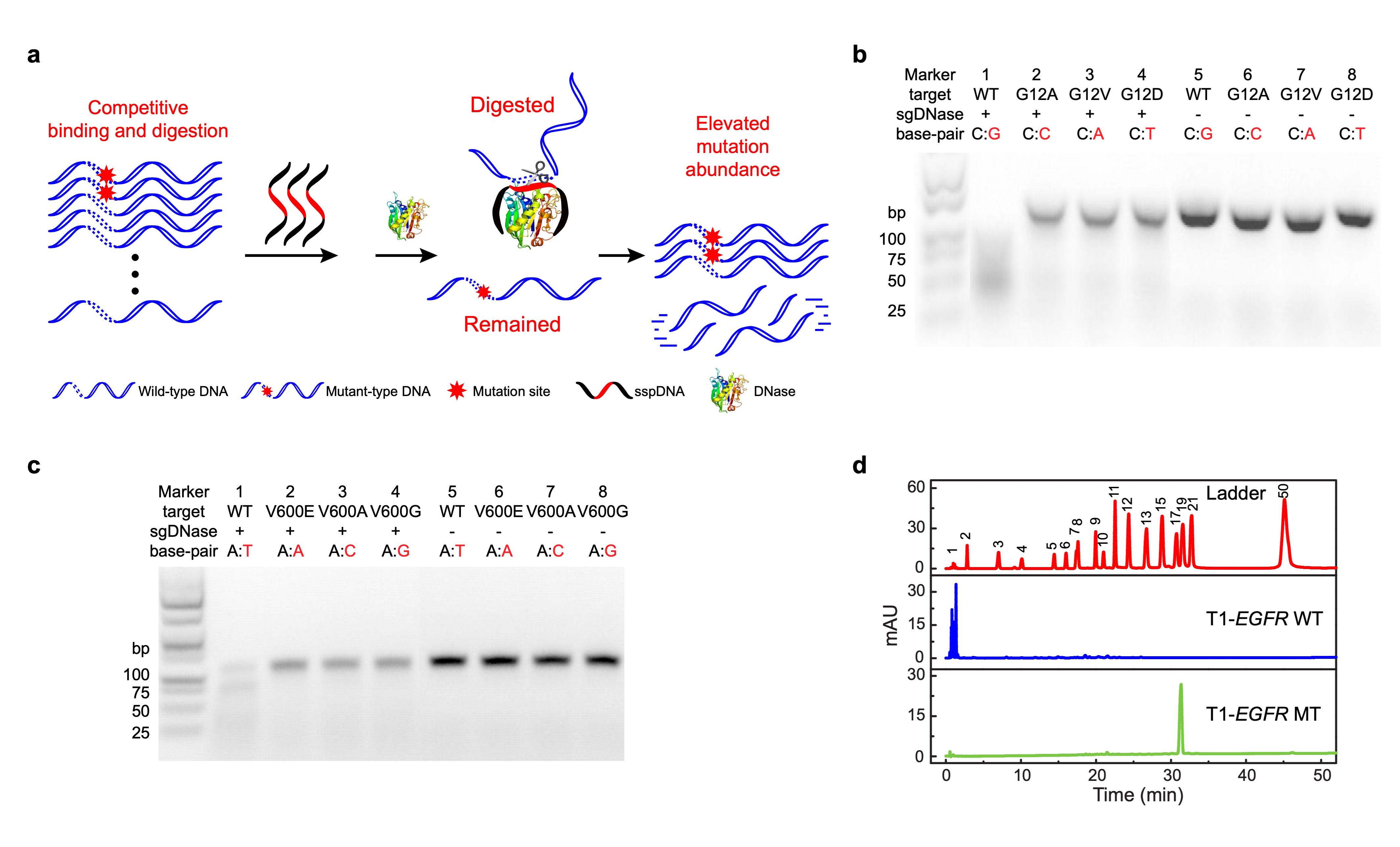
Fig. 1 | Proof of concept: phosphorothioated DNA guided deoxyribonuclease I (sgDNase). a, Illustration of the sgDNase system via a competitive binding and digestion mechanism. b-d, Determination of the single-base differentiation ability of sgDNase between fully matched wild-type strand and mismatched mutant-type strands with different types of mismatched bases using (b-c) agarose gel electrophoresis analysis and (d) HPLC of the digestion products of (b) KRAS G12 targets, (c) BRAF V600 targets and (d) EGFR L858 targets.
Enrichment of Mutations:
The effectiveness of sgDNase was demonstrated by enriching low-level mutant DNA strands, particularly the EGFR L858R mutation. By pretreating synthetic samples, we significantly increased the variant allele frequency (VAF) from 0.01% to 22% (Fig. 2a). Moreover, sgDNase was successful in enriching 18 different mutations with initial VAFs ranging from 0.01% to 10% (Fig. 2b). Furthermore, the sgDNase system enabled the simultaneous enrichment of different mutation types within mutational hotspots, such as KRAS G12 and G13, facilitating the quick identification of potential rare and unknown mutations (Fig. 2c).

Fig. 2 | Enrichment of single and multiplex hotspot mutations by sgDNase pretreatment. a, Comparison of Sanger sequencing chromatograms of synthetic EGFR L858R samples with different pre-VAFs (0% - 50%) before and after sgDNase pretreatment. b, Post-VAFs (measured by Sanger sequencing) of 18 clinically relevant target mutations with pre-VAFs varying from 0.01% to 10% after single-plexed enrichment by sgDNase. c, Next-generation sequencing (NGS) measurement results of the abundance of KRAS G12G13 mutations in the sample with a pre-VAF at 0.01% before and after sgDNase pretreatment. Variant to noise plots derived from sequencing data are depicted.
Direct Genomic DNA Pretreatment:
We extended the application of sgDNase to directly pretreat double-stranded genomic DNA using the double phosphorothioated DNA guided deoxyribonuclease I (dsgDNase) system. This approach significantly increased VAFs for various mutations, enabling their detection using multiple mutation detection methods (Fig. 3a). Notably, the sgDNase pretreatment allowed for accurate identification of ultra-low-abundance mutations in clinical biopsy samples (Fig. 3b and 3c).
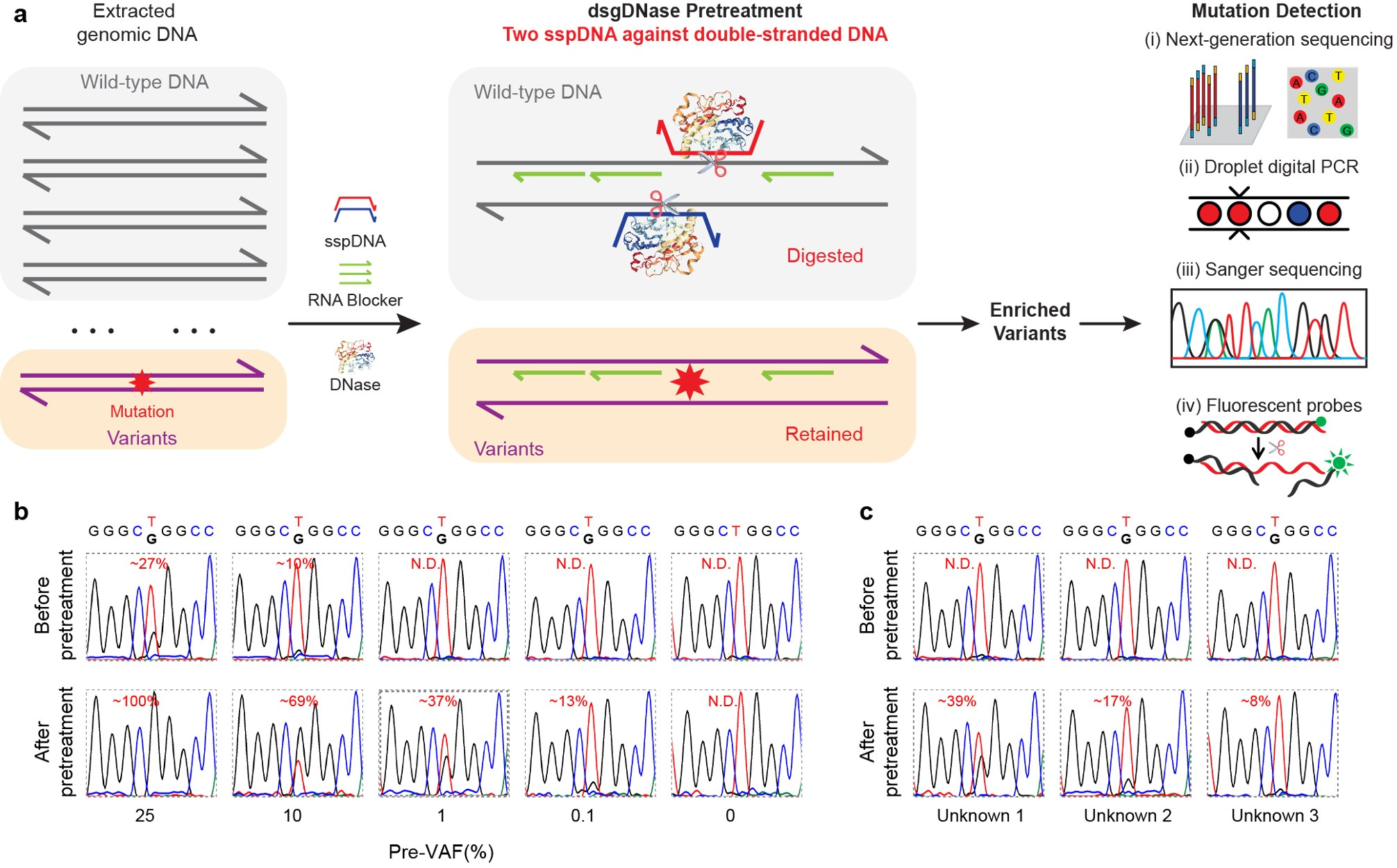
Fig. 3 | Direct pretreatment of genomic DNA by double sspDNA guided DNase (dsgDNase). a, Schematic illustration of direct enrichment of mutations in double-stranded genomic DNA by double phosphorothioated DNA guided deoxyribonuclease I (dsgDNase) system. b, Comparison of the Sanger sequencing results of genomic DNA with EGFR L858R mutation at different pre-VAFs (0% - 25%) before and after dsgDNase pretreatment. c, Sanger sequencing chromatograms of three unknown genomic DNA samples with EGFR L858R mutation before and after dsgDNase pretreatment.
Clinical implementation:
The sgDNase assay has been applied to more than 70 clinical samples and totally 16 positive samples were identified. As shown in Fig. 4a-b, without sgDNase pretreatment, only two tissue samples (Patients P2 and P5) were observed to be EGFR L858R mutation positive, while no positive blood samples were identified. After sgDNase pretreatment, one more tissue sample (Patient P7) and two blood samples (Patients P2 and P10) were identified as L858R mutation positive (Fig. 4a). The positive samples were further validated by NGS. Elevated VAFs were achieved in clinical samples regardless of sample types (blood, tissue) and disease types (lung cancer, colorectal cancer and venous malformations) after sgDNase pretreatment. sgDNase exhibited a clinical sensitivity of 94.74% and a clinical specificity of 98.58%.

Fig. 4 | Detection of ultralow-abundance mutations in clinical biopsy samples with sgDNase pretreatment. a, sgDNase applied to sixteen cfDNA and genomic DNA samples from eight NSCLC patients (P1-P8), four colorectal cancer patients (P9-P12) and standard reference DNA (SRD). P, T and B in the sample name represent patient, tissue sample (genomic DNA) and blood sample (cfDNA) respectively. b, Comparison of Sanger sequencing chromatograms of the EGFR L858R mutation in gDNA (tissue) and cfDNA (blood) samples from an NSCLC patient and a colorectal cancer patient before and after sgDNase pretreatment. c, Summary of the post-VAFs of cancer patients and negative gDNA control after sgDNase pretreatment (measured by Sanger sequencing). P value = 1.4E-76, from two-tailed Welch’s unpaired t-tests.
Implications for Precise Cancer Therapy:
The high-resolution capability, universality, ease of implementation, and robust enrichment capability of sgDNase hold significant potential for continuous mutation status assessment and precise cancer therapy. Additionally, sgDNase can be employed to selectively digest unwanted DNA sequences for various detection purposes, further enhancing its versatility and applicability.
Conclusion:
The development of the phosphorothioated DNA-guided endonuclease excision tool represents a groundbreaking advancement in DNA mutation detection. Its ability to remove wild-type DNA background and enrich low-frequency mutations opens up new possibilities for accurate diagnosis and targeted cancer therapy. The simplicity, efficiency, and broad applicability of this method make it a promising asset for the field of molecular diagnostics.
References:
- Vogelstein, B. et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 319, 525-532 (1988).
- Abascal, F. et al. Somatic mutation landscapes at single-molecule resolution. Nature (2021).
- Bustin, S.A. et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 55, 611-622 (2009).
- Baker, M. Digital PCR hits its stride. Nat. Methods 9, 541-544 (2012).
- Metzker, M.L. Sequencing technologies — the next generation. Nat. Rev. Genet. 11, 31-46 (2010).
- Xiao, X., Wu, T., Gu, F. & Zhao, M. Generation of artificial sequence-specific nucleases via a preassembled inert-template. Chem. Sci. 7, 2051-2057 (2016).
Follow the Topic
-
Nature Biomedical Engineering

This journal aspires to become the most prominent publishing venue in biomedical engineering by bringing together the most important advances in the discipline, enhancing their visibility, and providing overviews of the state of the art in each field.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
Full-text access to the paper :
https://rdcu.be/dhUlL