Targeting KRAS promoter G-quadruplex for KRAS oncogene repression: Structural basis of small molecule recognition of KRAS G-quadruplex
Published in Chemistry

KRAS is well-known as one of the most highly mutated oncogenes in human cancers and implicated in poor survival, particularly in pancreatic (~95%), colorectal (~41%), and lung (~32%) cancers.1,2 Notably, although KRAS is a highly pursued therapeutic target, direct inhibition of mutant KRAS oncoprotein has proven to be challenging in precision oncology for nearly 40 years.1 The KRAS oncoprotein has thus been considered as "undruggable". Therefore, it is essential to develop alternative KRAS signaling inhibitors.
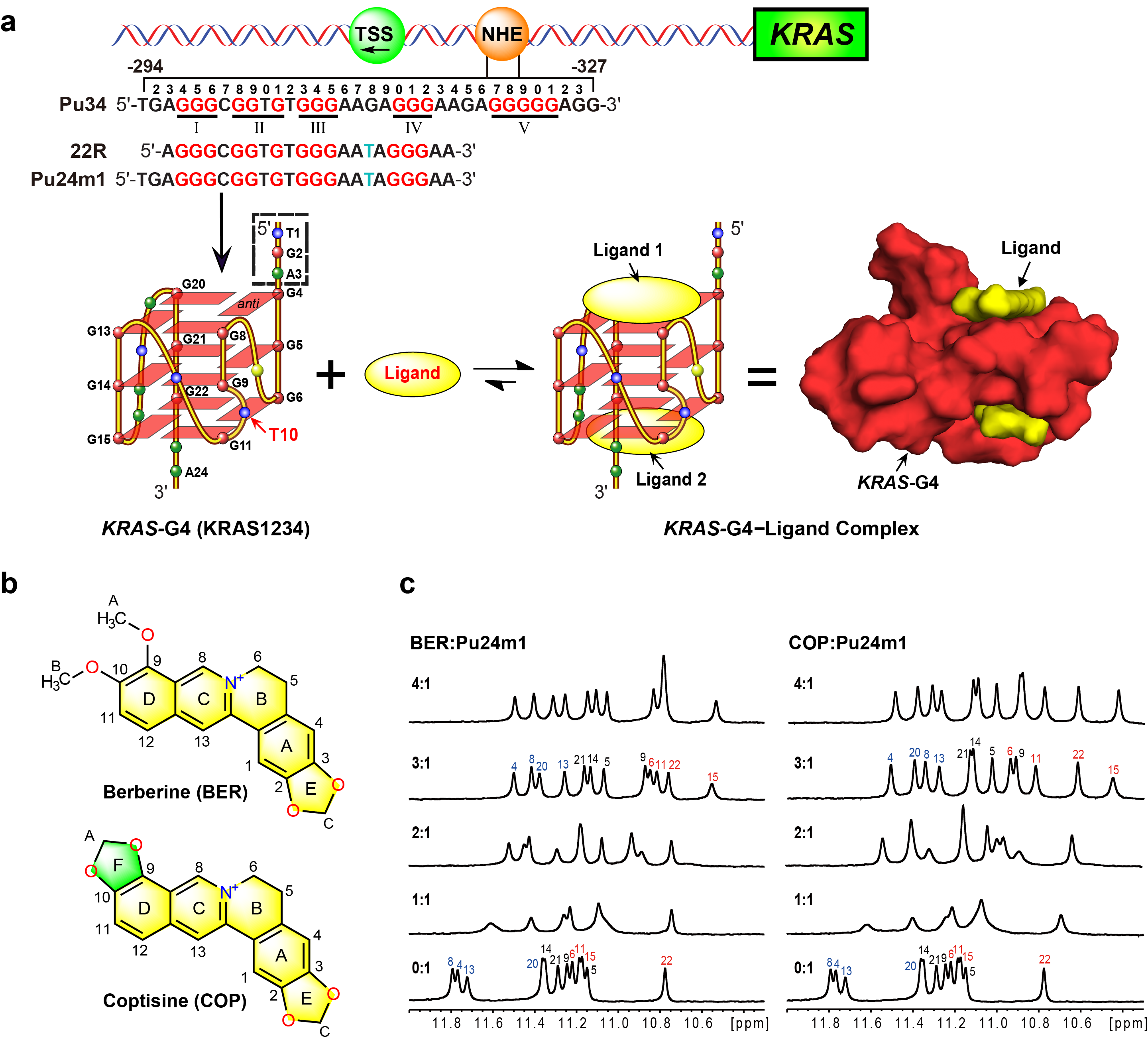
Figure 1. KRAS-G4 and its interaction with berberine and coptisine. (a) Schematic of the human KRAS gene promoter and the formation of a KRAS-G4 as well as its complex with small molecules (ligands). The G4-forming region of the NHE sequence is shown. The G-tracts implicated in KRAS-G4 formation are marked in red and mutations in cyan, respectively. (b) Chemical structures of berberine and coptisine with numbering. (c) 1D 1H NMR titration of Pu24m1 DNA with berberine and coptisine, respectively, with complete imino proton assignment.
The promoter region of the human KRAS oncogene has a critical GC-rich nuclease hypersensitive element, which can form a DNA G4 structure (KRAS-G4, Figure 1).3 KRAS-G4 has proved to be a transcriptional modulator, which is amenable to small-molecule targeting.4-6 Several free KRAS-G4 structures have been determined and a number of KRAS-G4 interactive small molecules (ligands) have been reported. However, no available KRAS-G4-ligand complex structure has yet been determined, which seriously hinders the structure-based rational design of KRAS-G4 targeting drugs.
We start to seek suitable KRAS-G4 binding ligands and to determine their complex structures back into 2020 when I just joined China Pharmaceutical University and established my lab with the help of Prof. Ling-Yi Kong.
As published free KRAS-G4 structure only has an adenine residue for the 5′-end flanking site, we think that is not enough for specific ligand recognition. We thus checked the genome sequence of the KRAS oncogene and chose the Pu24m1 DNA sequence which contains the wild-type TGA residues in the 5´-flanking site so that can better mimic the wild-type KRAS-G4. We then determined the NMR solution structure of free KRAS-G4 using Pu24m1 DNA sequence. With the obtained free KRAS-G4 structure, we screened several natural alkaloids and found that berberine (BER) and coptisine (COP) could strongly bind and stabilize the KRAS-G4 by using various biophysical experiments. We subsequently determined the high-resolution NMR solution structures of KRAS-G4 in complex with berberine and coptisine, respectively. The determined complex structures elucidate the detailed features for the specific recognition of the KRAS-G4 by small molecules. Moreover, berberine and coptisine significantly stalled the Taq DNA polymerase synthesis of the complementary strand DNAs and lowered the KRAS mRNA levels in cancer cells. Collectively, our study, for the first time, contributes structural insights into the ligand interactions with KRAS-G4 and provides a model system for the design of specific and potent KRAS-G4-interactive drugs.
References:
- Moore, A.R., Rosenberg, S.C., McCormick, F. & Malek, S. RAS-targeted therapies: is the undruggable drugged? Nature reviews. Drug discovery 19, 533-552 (2020).
- Buscail, L., Bournet, B. & Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nature reviews. Gastroenterology & hepatology 17, 153-168 (2020).
- Cogoi, S. & Xodo, L.E. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic acids research 34, 2536-2549 (2006).
- Cogoi, S., Paramasivam, M., Spolaore, B. & Xodo, L.E. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic acids research 36, 3765-3780 (2008).
- Amato, J., et al. HMGB1 binds to the KRAS promoter G-quadruplex: a new player in oncogene transcriptional regulation? Chemical communications (Cambridge, England) 54, 9442-9445 (2018).
- Cogoi, S., Ferino, A., Miglietta, G., Pedersen, E.B. & Xodo, L.E. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic acids research 46, 661-676 (2018).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in