The Hidden Faces of Tick Immune Cells: Navigating Fitness and Microbial Challenges
Published in Protocols & Methods, Cell & Molecular Biology, and Immunology

Ticks are ancient arthropods that transmit pathogens affecting both humans and animals. Unique characteristics such as their extended lifespan, exclusive reliance on blood meals throughout all life stages, ability to adapt to various environments, and exposure to diverse microorganisms make ticks unique compared with other arthropods. Despite these distinctive features, our understanding of arthropod biology is predominantly derived from model organisms like flies or mosquitoes, which represent only a fraction of the species found in nature.
Hemocytes are specialized arthropod immune cells that play a pivotal role in determining infection outcomes. Historically, tick immune cells have been categorized based on their cellular morphology. While this classification is useful, it remains incomplete as the ontogeny, plasticity, molecular features, and function of hemocytes during hematophagy and infection are still ill-defined. Therefore, we employed and developed advanced techniques to characterize the immune cells of the tick Ixodes scapularis, which transmits the Lyme disease spirochete Borrelia burgdorferi and the rickettsial agent Anaplasma phagocytophilum.
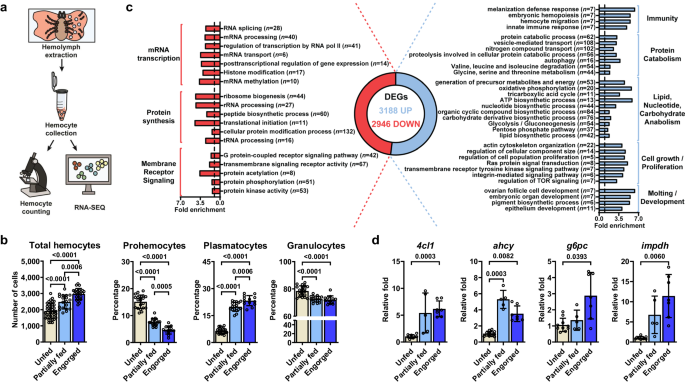
Initially, we refined a procedure for harvesting hemocytes from I. scapularis nymphs, a clinically relevant stage. This process revealed three commonly reported morphotypes: prohemocytes, plasmatocytes, and granulocytes. Surprisingly, an elevated total hemocyte count was observed following blood-feeding, particularly in the percentage of plasmatocytes. This observation suggests that hemocytes respond to metabolic changes induced by hematophagy.
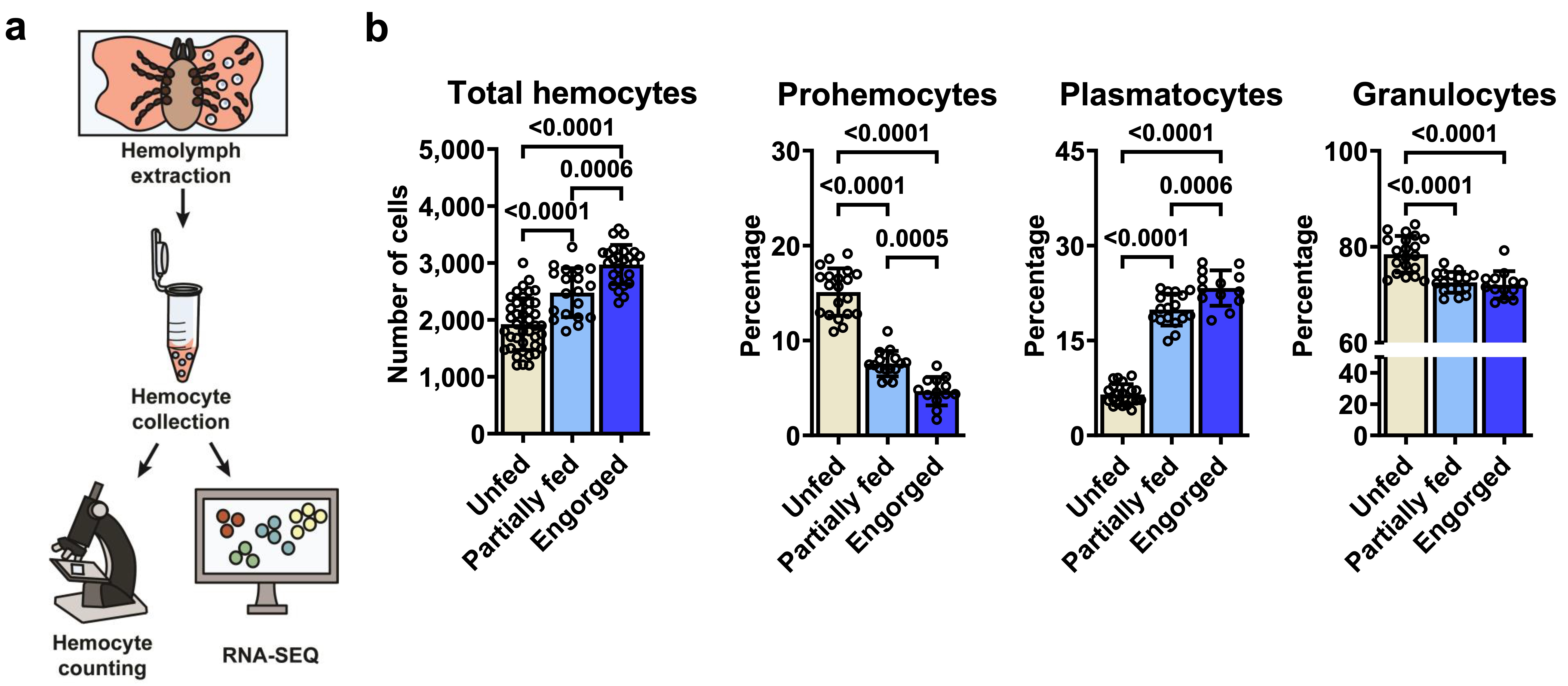
I. scapularis hemocytes undergo changes upon blood-feeding. (a) A schematic depiction of the hemocyte collection process. (b) The total hemocyte count and the proportions of distinct morphotypes are displayed for unfed (ivory), partially fed (light blue), or engorged (dark blue) nymphs.
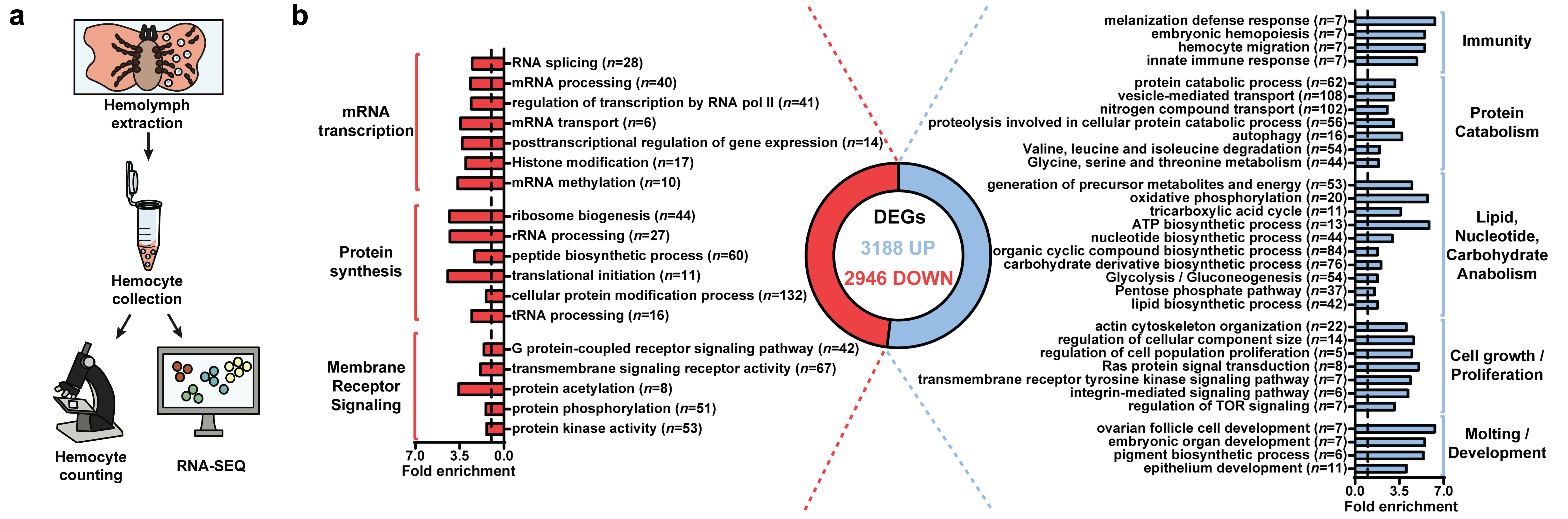
Our subsequent goal was to explore the global transcriptional profile of hemocytes activated by a blood meal. Our analysis revealed distinct patterns linked to immunity, metabolism, cell proliferation/growth, and arthropod molting/development. Consequently, the genetic program of I. scapularis immune cells proves to be dynamic during hematophagy, encompassing functions that extend beyond mere immunity.
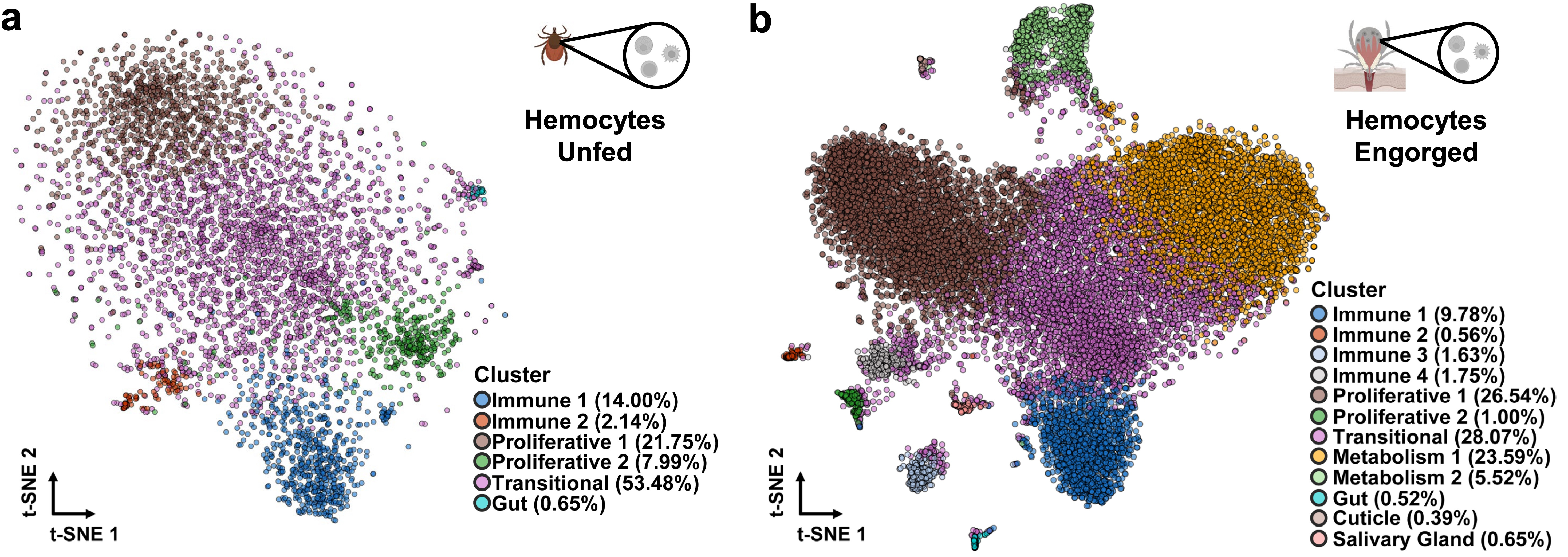
Next, we investigated whether the generalized transcriptional changes reflected variations among specific cells by profiling individual hemocytes. This analysis unveiled several distinct clusters during both hematophagy and infection. Additionally, we identified molecular markers for each hemocyte subtype and predicted their ontogeny. Notably, our findings suggest the existence of an oligopotent subpopulation that undergoes differentiation into more specialized subtypes associated with immune and metabolic functions, a phenomenon triggered by hematophagy.
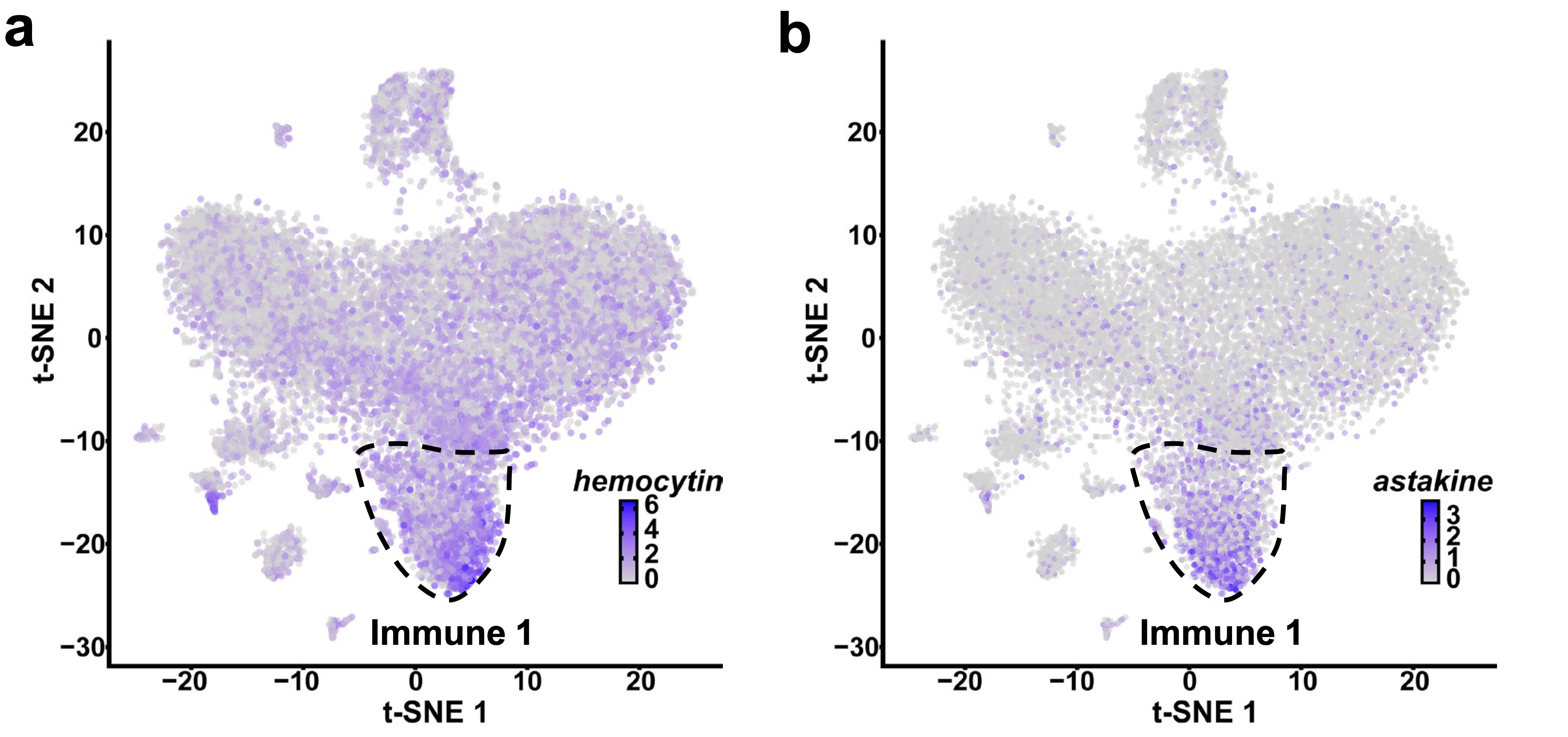
Then, we assessed the impact of bacterial infection and found specific subpopulations or genes that were differentially modulated in response to A. phagocytophilum or B. burgdorferi. Particularly, the Immune 1 hemocyte cluster, which expresses hemocytin, astakine, and genes related to phagocytosis, represented a subpopulation of cells that responded to infection in I. scapularis. Therefore, our focus shifted to this cluster and its marker genes.
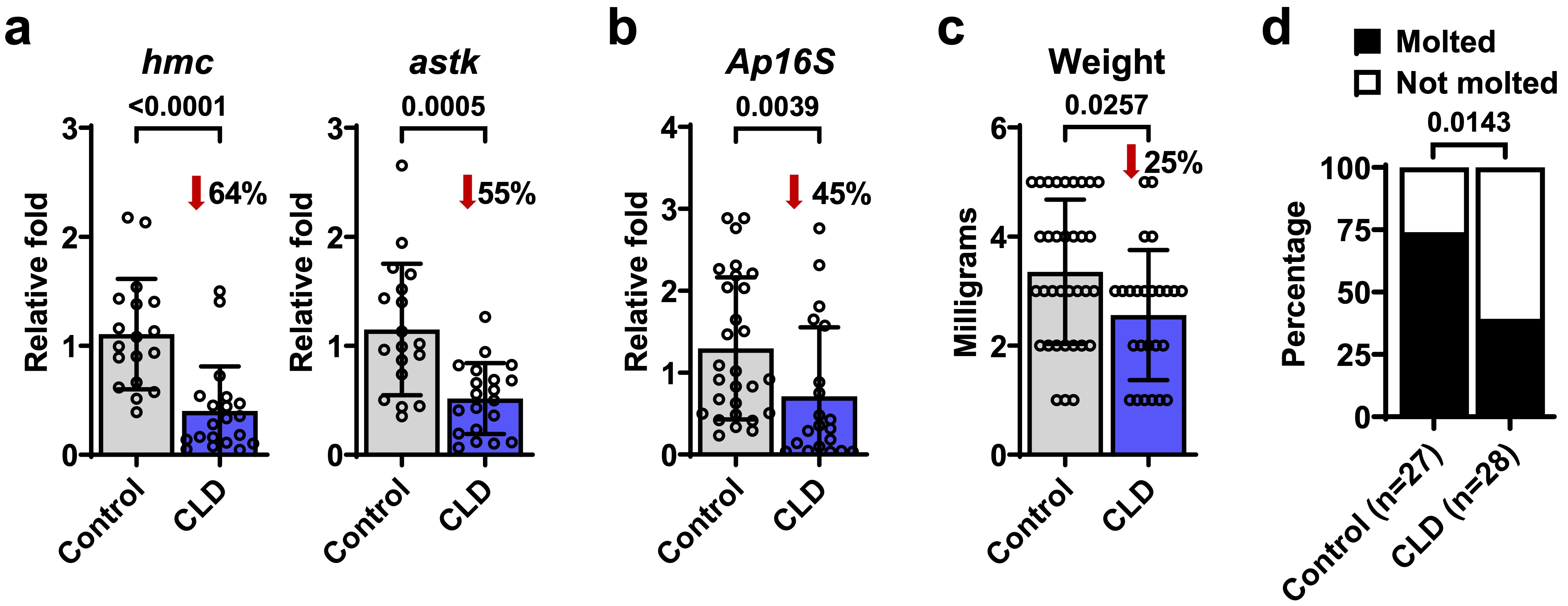
Clodronate liposomes (CLD) were employed to deplete phagocytic hemocytes, leading to a significant decrease in the proportion of prohemocytes and plasmatocytes, accompanied by a reduction in the expression levels of both hemocytin and astakine, supporting the association of phagocytic activity with the Immune 1 cluster. Intriguingly, CLD treatment also resulted in reduced loads of A. phagocytophilum, diminished tick weight, and impaired molting success. These findings suggest that phagocytic hemocytes have pleiotropic effects on tick immunity, feeding, and ecdysis in I. scapularis.
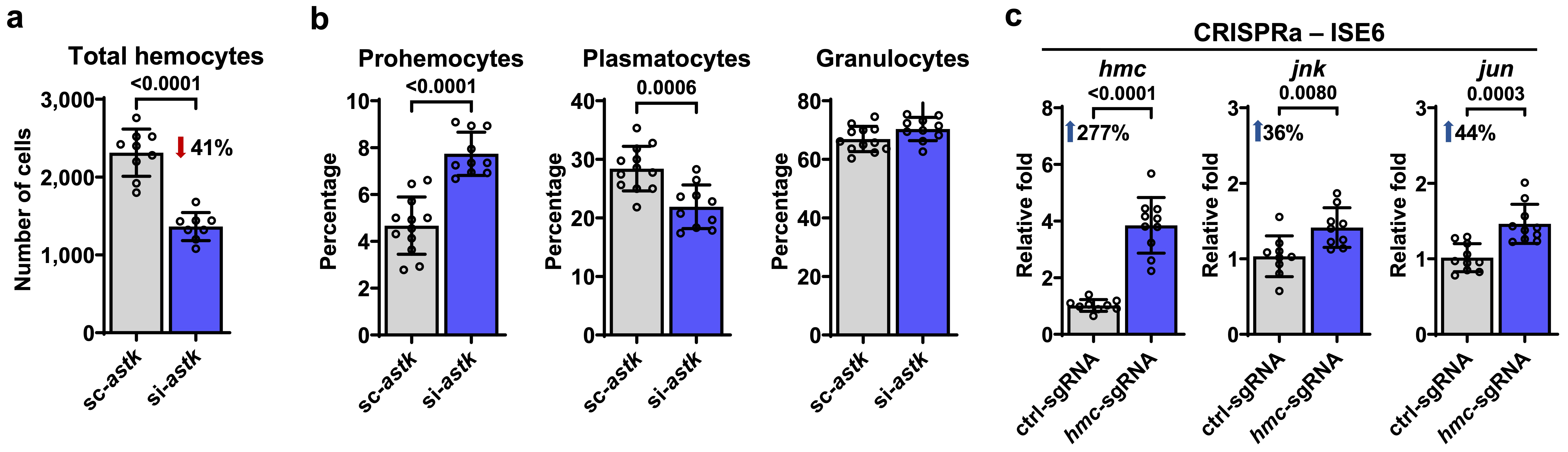
Finally, we functionally characterized the roles of the marker genes astakine and hemocytin using small interfering RNA (siRNA) and CRISPR activation (CRISPRa), which respectively reduce or increase the expression of targeted genes. Our results demonstrated that both genes play crucial roles in bacterial acquisition, as well as tick feeding and ecdysis. Particularly, astakine was found to influence hemocyte proliferation, while hemocytin positively regulates the c-jun n-terminal kinase (JNK) pathway.
Through a meticulous examination of hemocytes at a single-cell resolution, combined with complementary methodologies, we uncovered the heterogeneity of immune cells in ticks, with roles that extend beyond the traditional response to infection. Notably, hemocytes enriched in genes associated with metabolic functions driven by hematophagy, indicate involvement in the feeding process by metabolizing nutrients and xenobiotics present in the blood meal.
Additionally, immune-related hemocytes not only exhibited an overrepresentation of genes involved in antimicrobial responses but also in tissue remodeling, a crucial process during blood meal acquisition. Considering the established immunological roles of plasmatocytes and granulocytes in ticks, we hypothesize that these immune clusters encompass these morphotypes.
Furthermore, our investigation identified hemocytes specialized in cellular proliferation, including cells expressing genes related to ecdysteroid biosynthesis, essential for regulating molting and development. We categorize these proliferative clusters as prohemocytes, given the stem cell-like properties of this morphotype.
In summary, we leveraged the synergy of systems biology and reductionist methods to uncover unexpected roles of tick immune cells. The extensive hemocyte dataset presented in this study, along with the identification of cell type-specific marker genes and experimental tools, constitutes an asset for comparative biology. Undoubtedly, this research will be instrumental in advancing future mechanistic studies in ticks, with the potential to make significant contributions to ongoing efforts in combating vector-borne diseases.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Ask the Editor - Immunology, Pathogenesis, Inflammation and Innate Immunity
Got a question for the editor about the complement system in health and disease? Ask it here!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in