The successful transaction between wheat and transposons
Published in Ecology & Evolution

Curiosity at the beginning: why wheat has high plasticity?
Wheat is one of the most common staple foods in the world. Most of the wheat we see today is hexaploid, having undergone two rounds of hybridization and polyploidization events between three diploid ancestors that adapt to different environments. Due to the merge of the three genomes, common wheat has three synergistic and divergent subgenomes, which may allow it to spread rapidly around the world with high environmental plasticity compared to its diploid ancestors. Since the release of high-quality hexaploid wheat genome assembly by the International Wheat Genome Sequencing Consortium (IWGSC) in 2018, our team invested a lot of effort over the years, focusing on subgenome-diversified and -coordinated regulation from the following points:
- Demonstrated subgenome-dominant TE families associated with maintaining subgenome-specific chromatin territories in common wheat[1].
- Constructed the epigenomic profile of common wheat and identified regulatory cis-elements on a genome-wide scale[2].
- Detected subgenome asymmetric regulation in common wheat at different growth and developmental stages[3].
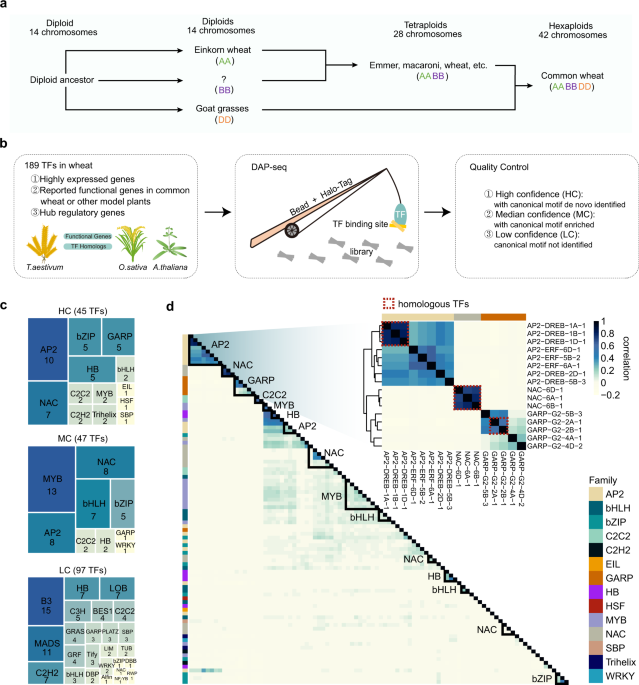
- Profiled the bindings of hundreds of transcription factors (TFs) and demonstrated lineage-specific TEs rewired the wheat transcriptional regulatory network[4].
The power of TEs should not be underestimated
It was not our original intention to focus on the contribution of TEs to the wheat genome evolution and adaptation. But as our research progressed, many of the "whys" were deposited on TEs. To maintain genomic stability, TEs are generally repressed by DNA methylation, with low chromatin accessibility. However, there are also exceptional cases showing that TEs can be involved in regulatory networks. Most Triticeae species, including Secale cereale, Hordeum vulgare, diploid, tetraploid and hexaploid wheat, have abundant TEs in their genome (~80%), and TEs may make a specific contribution to genome evolution and have some important implications for the adaptation of Triticeae species. Based on our results mentioned above, we began to focus on how TEs that contribute to sequence diversity affect the regulatory plasticity among subgenomes, which may result in the high environmental adaptability of common wheat.
The expansion of paleo- and neo-TEs contributes to the high plasticity of wheat
In our paper, we obtained the binding data for 189 functionally important TFs using DAP-seq. We found that subgenome-specific TFBSs share significant sequence similarity within subgenomes and are associated with subgenome-specific TE expansions, suggesting that TEs specifically expanded in diploid progenitors contribute to subgenome-specific regulatory expansion.
Intriguingly, despite the intergenic regions are nearly turned over by TEs and are highly divergent among subgenomes, TEs also contribute to synergistic transcription among subgenomes. We found that in the triads with balanced TFBSs among three subgenomes, TE-TFBS mostly presents in only one subgenome, raising an interesting question of why subgenome-unbalanced TE insertion is accompanied by the balanced TF binding. The answer is that the wheat genome has undergone rounds of TEs bursts and degeneration, but some internal TFBSs were subject to parallel selection and were highly conserved across subgenomes. As a result, some TEs no longer had the classical TE structure and only the TFBSs were preserved. By tracing the insertion time of these decayed TEs, we observed that paleo-expansion occurred in the common ancestor of diploid wheat (about 5 million years ago). The above results suggest that the ancient expansion of TEs contributes to the convergent regulation in common wheat.
In summary, we discovered the contributions of ancient and recent TE expansions to the regulation of subgenome divergence and convergence in common wheat, and demonstrated how the plasticity of TE repertoires affects the plasticity of polyploidy regulation.
For details, please refer to our paper at https://www.nature.com/articles/s41467-022-34290-w.
Photo: Yilin Xie
References:
- Jia, J., et al., Homology-mediated inter-chromosomal interactions in hexaploid wheat lead to specific subgenome territories following polyploidization and introgression. Genome Biol, 2021. 22(1): p. 26.
- Li, Z., et al., The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements. Genome Biol, 2019. 20(1): p. 139.
- Wang, M., et al., An atlas of wheat epigenetic regulatory elements reveals subgenome divergence in the regulation of development and stress responses. Plant Cell, 2021. 33(4): p. 865-881.
- Zhang, Y., et al., Evolutionary rewiring of the wheat transcriptional regulatory network by lineage-specific transposable elements. Genome Res, 2021.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in