The Traffic Lights in Wheat’s Adaptive Evolution: Epigenetic Buffering System
Published in Genetics & Genomics and Plant Science

Allopolyploidization, encompassing hybridization and genome doubling events, fixes hybrid vigor and is a major driver of genome evolution and adaptation. However, the merger of different genomes may result in genomic conflicts, typical examples include competition between parental genomes and the rapid loss or repression of homoeologous gene copies, raising a major question regarding how genetic heterogeneity is interpreted and regulated to enable environmental plasticity in polyploids.
Our research group has been committed to revealing how subgenomic diversity contributes to polyploidy regulation from the perspectives of chromatin 3D structure [1], histone modifications [2,3], transcription factor binding [4,5] and non-coding regulatory elements transcription [6]. However, the causal relationship between subgenomic divergent regulation and phenotypic plasticity remains unclear.
LHP1-mediated buffering of subgenome diversity confers developmental stability and environmental adaptability
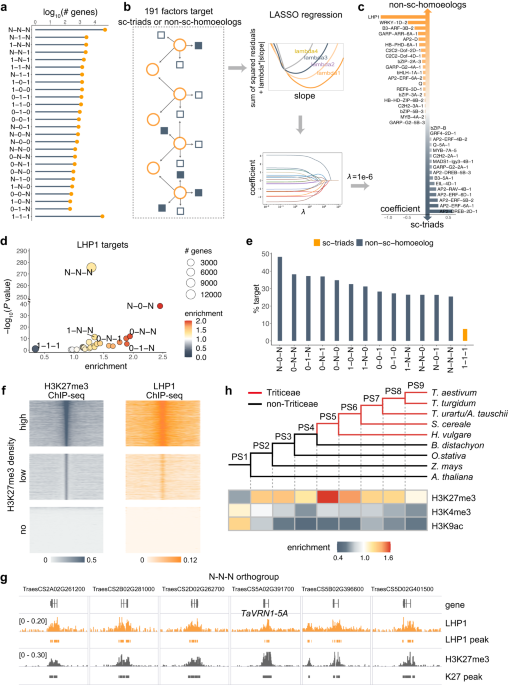
Identifying the factors regulating subgenome variations is vital for elucidating the causal mechanism underlying polyploid adaptation and evolution. We used machine learning approach based on the genome-wide binding of 191 sets of transcription factors and epigenetic factors previously published [5] and identified LHP1 as a key factor in regulating subgenome diversity. LHP1 is a member of the Polycomb family of proteins (PcGs) and is responsible for recognizing and spreading the repressive histone modification H3K27me3.We next knock out the LHP1 homeologous genes and profiled the transcriptomic and epigenomic map of the mutants to determine LHP1 targets. The target genes are generally induced in mutants, particularly in the subgenome divergent genes involved disease resistance.
To investigate which types of pathogen responses LHP1 preferentially regulates, we collected publicly available transcriptomic data related to various pathogen responses. We found that deletion of LHP1 resulted in similar transcriptome changes with introduction of a wheat stripe rust (Pst) resistance locus. Pst infection suggested LHP1 mutation conferred apparent stripe rust resistance to wheat seedlings. We further demonstrated that Pst infection eliminates the inhibitory effect of LHP1 on subgenomically diverse defense genes, releasing pre-existing potential subgenomic variation.
LHP1-mediated epigenetic buffering system regulates adaptive evolution
Increasing evidence suggests that the rate of evolution of a gene is primarily influenced by its expression level rather than its functional importance. The strength and breadth of LHP1 repression of subgenomically diverse genes suggests that LHP1 may influence evolutionary processes. We examined the relationship between LHP1 binding and target sequence variation at the population level. There are two main sources of subgenomic diversity in common wheat: acquired from diploid ancestors (i.e., variation already present in diploids) and newly generated in polyploid populations. We first compared these two types variants in a pairwise framework, which were highly correlated. Regions of differentiation between diploid ancestors tend to further diverge after allopolyploidization, representing neutral or weakly deleterious loci that may arise due to genetic drift. These regions are enriched for LHP1 binding and H3K27me3 modification. High correlation between LHP1 binding, H3K27me3 modification and population diversity were detected on genome-wide scale. Therefore, LHP1-mediated H3K27me3 modification may promote neutral variation and protect diversifying genes from strong purifying selection.
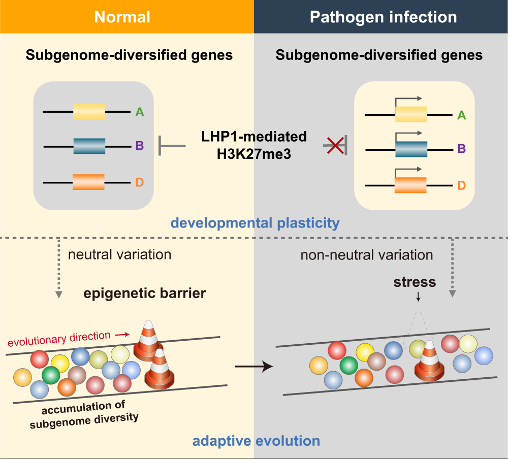
Model illustrating the LHP1-mediated conditional repression of subgenome-diversified defenses, which leads to developmental plasticity and facilitates adaptive evolution in allopolyploid wheat.
In summary, this study discovered LHP1 as a key factor regulating subgenomic divergence and disease resistance genes by quantitatively integrating multi-omics data via a machine learning scheme and genetic validations. Under normal conditions, LHP1-mediated epigenetic buffering system protects normal development from environmental changes and leads to the variation accumulation between subgenomes. Pathogen infection eliminated LHP1 repression, enabling previously unavailable phenotypic variants to surface, thereby releasing defense cascades and facilitating the timely fixation of favorable mutations in particular environmental niches.
Hint: improving agronomic traits by epigenetic manipulation
From a genetic improvement perspective, this work demonstrates that latent genetic diversity can be unlocked by manipulating epigenetic systems. Common wheat is thought to lack sufficient diversity to cope with environmental stress due to polyploidy bottlenecks, so introducing resistance genes or loci from wild relatives into modern varieties has been the main strategy. The wheat genome contains a large number of inactive or silent gene loci that are highly divergent among subgenomes, some of which are associated with key agronomic traits. If properly activated, they are expected to improve relevant agronomic traits. In this study, although the wheat variety "JW1" was highly susceptible to stripe rust, apparent stripe rust resistance could be conferred on seedlings by manipulating the epigenetic system. It shows that mining and utilizing gene resources hidden in the wheat genome is a new potential genetic strategy to improve wheat agronomic traits.
The prerequisite for epigenetic manipulation is the elucidation of specific mechanisms. In previous work, through cooperation with multiple laboratories, we demonstrated the specific factors and components of plant PcG involved in environmental perception and response by integrative data analysis [7-11], which may provide theoretical or technical clues.
For details, please refer to our paper at https://www.nature.com/articles/s41467-023-43178-2
References:
1. Homology-mediated inter-chromosomal interactions in hexaploid wheat lead to specific subgenome territories following polyploidization and introgression (2021) Genome Biology, 22(1):26
2. The bread wheat epigenomic map reveals distinct chromatin architectural and evolutionary features of functional genetic elements (2019) Genome Biology, 20(1):139
3. An atlas of wheat epigenetic regulatory elements reveals subgenome-divergence in the regulation of development and stress responses (2021) The Plant Cell, 33(4):865-881
4. Evolutionary rewiring of the wheat transcriptional regulatory network by lineage-specific transposable elements (2021) Genome Research, 31(12):2276-2289
5. Transposable elements orchestrate subgenome-convergent and -divergent transcription in common wheat (2022) Nature Communications, 13(1):6940
6. Transposable element-initiated enhancer-like elements generate the subgenome-biased spike specificity of polyploid wheat (2023) Nature Communications. 2023, 14, Article number: 7465
7. Arabidopsis flower and embryo developmental genes are repressed in seedlings by different combinations of Polycomb group proteins in association with distinct sets of cis-regulatory elements (2018) PLoS Genetics, 12(1): e1005771.
8. Polycomb repressive complex 2 attenuates ABA-induced senescence in Arabidopsis (2019) The Plant Journal, 97(2):368-377
9. Telobox motifs recruit CLF/SWN-PRC2 for H3K27me3 deposition via TRB factors in Arabidopsis (2018), Nature Genetics, 50(5):638-644
10. Enhanced sustainable green revolution yield via nitrogen-responsive chromatin modulation in rice (2020) Science, 367(6478):eaaz2046
11. Glucose-driven TOR-FIE-PRC2 signaling controls plant development (2022) Nature, 609(7929):986-993
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in