
Why study Atlantic salmon? Because as far as fish go, they are as important and iconic as it gets. These fish have huge commercial significance and also represent keystone species in natural ecosystems, making the conservation of wild populations of major societal value. Salmonids also have an interesting and complicated genome, having gone through a whole genome duplication quite recently (well… 95 million years ago!).

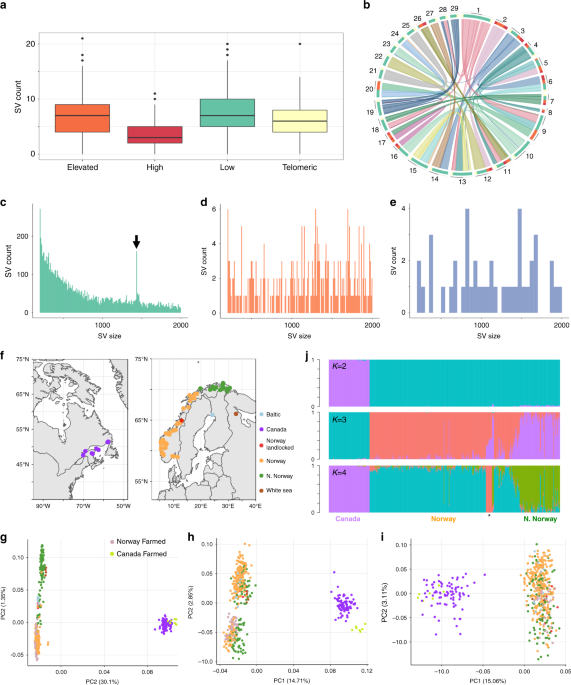
With over 5,000 studies published on Atlantic salmon, none had yet attempted to define structural variants (SVs, e.g. DNA deletions, duplications and inversions) across the genome. SVs can impact the function and regulation of genes, influencing normal phenotypic variation and being causative of a range of human diseases. Despite growing recognition of their importance, finding real SVs using widely available genome resequencing data remains (really!) difficult: there is a very high rate of false detection, meaning that distinguishing true calls from ‘bioinformatic garbage’ is challenging. This is one key reason why SVs are considerably understudied compared to SNPs.
.jpg)
In our study (click here), we set out to identify SVs in Atlantic salmon by resequencing the genomes of 492 fish from diverse wild and farmed populations. This was a large effort, involving many great international collaborators. The hardest part was finding real SVs. In a series of disappointing false calls and failed attempts at bioinformatically filtering out bad data, we realised no filtering step provided high enough accuracy. With the help of computer scientist, and human SV expert, Ryan Layer (CU Boulder) we added the crucial step of visualising the calls to retain only high confidence ones. To achieve our final dataset, we manually scored 165,116 different SV calls, 90% of which were dropped. A tool called SV-plaudit made the job efficient while working in a small team. We proved the accuracy of this visual validation step using long-read sequencing and hope it will help reduce the time-consuming and costly step of validating calls in the laboratory in future studies.
.jpg)
Once the above crucial work was achieved, we investigated the biological role of 15,843 novel variants in Atlantic salmon. We found that many overlapped coding genes, some with large, predicted effects and evidence that the accumulation of deletions was aided by ancestral genome duplication. We also identified a mobile element that may still be active in salmon genomes. Interestingly, many SVs were found within or near brain-expressed genes involved in behaviour. Farmed salmon populations have accumulated more of these alleles in their genome and were already known to show different behaviours to their wild relatives. These genes are linked to many neurological conditions in humans and we speculate that domestication selection on linked SVs has helped farmed fish to adapt behaviourally to aquaculture conditions.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in