Throughput is the key to the success of automated cell sorting
Published in Microbiology

In the paper, we demonstrate a complete pipeline to link microbial metabolic phenotypes to their genotypes. Within a complex natural microbial community, cells able to metabolize a particular compound can be labelled with deuterium (D) by incubating a community sample in D2O-containing medium supplemented by the unlabeled compound of interest. Our device uses an automated Raman-based microfluidic platform to sort the labelled cells for use in either DNA sequencing or cultivation. Given the potentially high diversity and variation in abundance of labelled cells in natural systems, as we demonstrate in the paper for the group of microbes with the ability to metabolize mucin in a mammalian gut microbe community, throughput will determine how many labelled taxa can be retrieved. This research was inspired by the need to provide an alternative to the extremely low throughput in manual Raman sorting. The Wagner (homepage) and Berry (homepage) groups in Vienna (coauthors on the paper) had been seeking to find a way to increase throughput of their manual Raman sorting technique of microbes with a function of interest that was published in PNAS in 20151, because this method had a very limited throughput (1–2 cells/h), and is laborious for the user. For glass capillary-based manual sorting1, (i) a user must fill an open-ended glass capillary (rectangular cross section; length ~10 cm) with the buffer medium followed by the sample, seal the ends to avoid evaporation over the next hours during sorting, then fix the capillary on the microscope stage – all with great care to avoid contamination. (ii) The user then manually searches for the single cells within the capillary using optical tweezers (by using a joystick that controls the xyz-position of the microscope stage). (iii) When a cell is captured, the user performs a Raman measurement to determine whether the cell is deuterium labelled or not. (iv) If the captured cell is labelled, it is carefully translocated to the sample-free end of the capillary. This requires it to be pulled with the optical tweezers against fluid drag without touching any walls, which would otherwise lead to immediate cell loss due to sticking. (v) The cell is released, the optical tweezers are moved back to the sample region, and a new sorting procedure starts over. (vi) After repeating the procedure for multiple labelled cells, the end of the capillary is broken off and placed in a sterile tube for the recovery of the sorted cells.

Fig. 1 | Microscope objective and capillary during manual cell sorting (image courtesy of Michael Wagner, David Berry, and Márton Palatinszky).
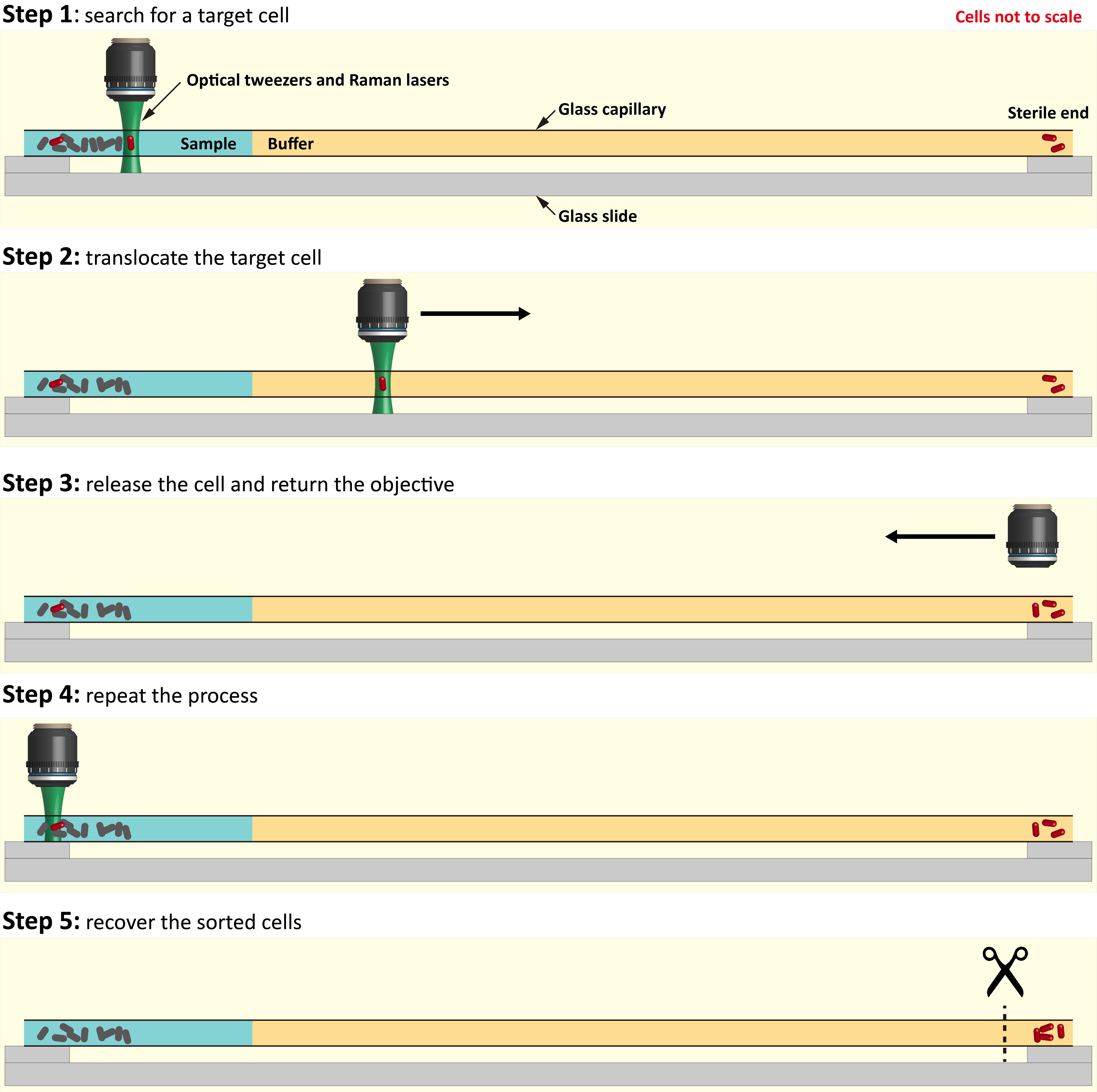
Fig. 2 | The manual cell sorting procedure. Our platform provides an automated procedure with around 100-fold higher throughput.
With the manual procedure, the Vienna researchers were able for the first time to sort deuterium labeled microbial cells from complex communities for subsequent DNA amplification and sequencing, but as described above only 1–2 cells/h could be sorted. First, it takes time to search for the labelled cells in the capillary. Second, given that a higher travelling speed of the optical tweezers increases the fluid drag on the captured cell, which leads to cell loss, the labelled cells can be translocated to the sample-free region at a speed of only ~500 µm/s. It thus takes 400 s (6.7 min) to move the cell to the sample-free region and release it, and to return the optical tweezers back to the sample region. Third, there are several other practical reasons that increase the probability that labelled cells are lost during the manual translocation between the ends of the capillary, further slowing the sorting speed.
Our platform provides throughput that is two orders of magnitude greater (200–500 cells/h) than the manual sorting methods described above, and thus makes function-based sorting of microbial taxa also feasible in many systems that were previously inaccessible. Sorted cells can be used for direct DNA amplification and sequencing (mini-metagenomics of cells with a specific function). Alternatively, the Raman-sorting device can be used as a front-end for subsequent FACS-sorting to perform single-cell genomics. For this purpose approx. 1,000 sorted cells per sample are required2. As an example, in a natural community in which 75% of the cells are labelled (active), our platform would enable the collection of sufficient cells in 2.6–6.7 hours of automated sorting (1,000 cells / (500 cells/h ×75%) to 1,000 cells / (200 cells/h ×75%)), whereas manual sorting would require 500–1,000 hours of sorting by a researcher (62.5–125 days; 1,000 cells / 2 cells/h to 1,000 cells / 1 cell/h). Using our method, the entire process is automated and requires no human intervention during the sorting, which drastically reduces the labour involved. Furthermore, our platform is – in contrast to other methods – perfectly suitable for subsequent cultivation of collected cells, because cells require no fixation before sorting, the laser exposure to the cell is short and the fluid flow reduces the laser-induced heating of the cell (which might otherwise lead to cell damage). By enabling both single-cell genomics and cultivation for further phenotypic characterization, our platform opens the door to studies to dissect the ecology of complex microbial communities and isolate strains with functions of interest.
Reference cited:
1. Berry, D. et al. Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. U.S.A. 112, E194-E203 (2015).
2. Kashtan, N. et al. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344, 416-420 (2014).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in