To bead or not to bead
Published in Microbiology

When my PhD student Manupriyam Dubey, David Johnson and myself started on the idea of a soil microbial interactome, we were intrigued by the question of the roles of interspecific interactions in microbiome development and functioning. At the time microbial species interactions were intensively studied in so-called range expansion experiments, in which lab strains are tagged with fluorescent colors, mixed and grown on agar plates. Starting from the center, expansion develops into beautiful patterns of mixed growth, from which interaction types can be deduced.
However, our focus was soil microbial communities, which are complex and highly diverse, with most species not in pure culture nor immediately amenable to fluorescent protein tagging. Paired range expansion experiments were thus clearly inappropriate for our purpose. We clearly needed a methodology that could follow growth interactions of natural species pairs, and that, given the high species diversity among soil bacteria, would be reasonably high-throughput. We figured if we could miniaturize ‘range expansion’, we could still rely on species growth to deduce interactions because many interactions are expected to develop during growth (e.g., competition for food, metabolite sharing).
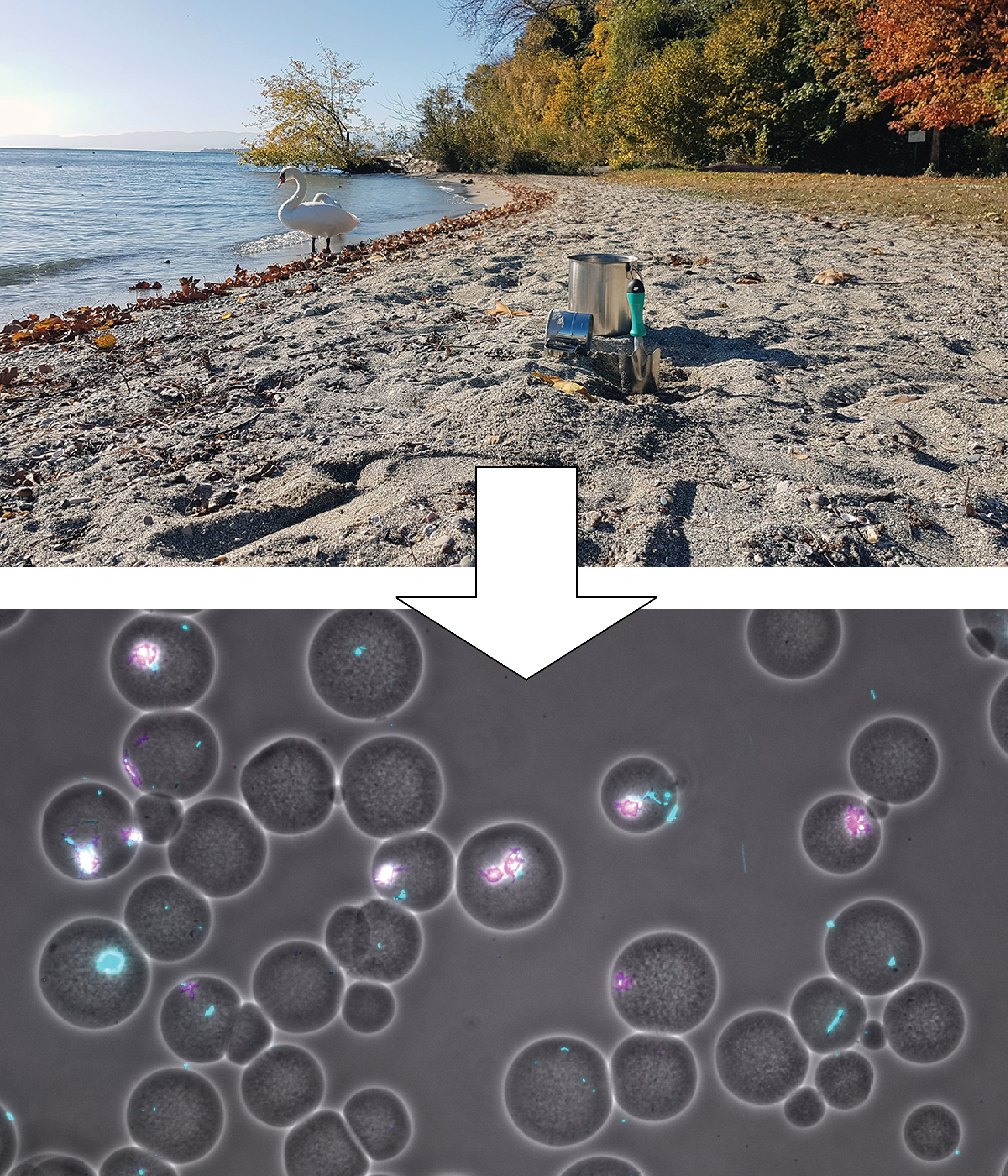
From previous work in the lab, we had experience with agarose mini-beads, and we felt that they could be used as appropriate containers to embed founder cell pairs, which we could then incubate under defined conditions. Cell division would result in the formation of small expanding colonies, which we could stain with a fluorescent dye and image. Indeed we managed to extract cells from a sandy soil from a Lake Geneva beach and produced millions of beads with random founder cells. We also observed plentiful growth, causing the need to reduce carbon amounts in the growth medium to a low level in order to prevent cells to burst from the beads.
Quantitative growth imaging, however, was a harder nut to crack. We teamed up with a specialist for (yeast) quantitative imaging within our Department (Serge Pelet), who wrote the Matlab script to find bead boundaries and quantify cells and their growth within beads. With help of Noushin Hadadi, a postdoc in the lab who was experienced in large data analysis, we expanded the script to go through hundreds and hundreds of images in automized fashion and extract relevant growth data. This all looked very promising and we obtained a lot of interesting data. However, our first submitted manuscript on this work was very poorly received and judged far too unfocused and complicated.
We went back to the drawing board to find and clarify the weak points in our study. One of the things we had noticed was that most of the (random) partnerships of the soil bacteria in the beads actually developed extremely unbalanced, with one partner producing a very small and the other a much bigger microcolony. This suggested on average a negative influence between pairs. As a result of the random cell encapsulation, however, many beads actually started with a single cell. And curiously, beads with single founder cells on average developed much poorer than pairs. This suggested that having a (random) partner is on average still beneficial for the growth of soil bacteria, perhaps because many of them display auxotrophies or other dependencies.
Growth in beads thus seemed to intensify interspecific interactions, perhaps as a result of the confined spatial environment and close distance between founding cells within beads. We then wondered whether ‘fragmented’ growth in confined small environments would have an influence on the diversity of the soil community as a whole. We thus contrasted community growth in beads (fragmented as random pairs and single founder cells) with regular mixed liquid suspensed growth. To our surprise, species diversity of the communities grown in completely suspended culture remained much higher than in the microbeads. This was in agreement with our idea that fragmented growth in small confined environments would aggravate the effect of interspecific interactions, causing imbalanced growth and apparent species loss.
To support our findings, we developed a model to simulate community growth of individual founder cells, random founder cell pairs, or representative mixtures of 200,000 cells in confined or mixed environments. We included and tested how different types of interspecific interactions influenced the growth and final community development. This largely confirmed the increased importance of interactions in paired confined growth but dissipating at mixed conditions.
We hypothesize that this effect of “environmental connectivity”, as we tried to formulate it, is relevant for many natural microbial communities that are characterized by spatially and/or temporarily confined microscale growth environments. On the other hand, the concept leads to a lot of questions. For example, the soil environment is actually characterized by multitudes of at least temporarily fragmented small growth environments, suggesting confined bead growth to be a good approximation. But if this would cause diversity loss, then how can it be that soil microbiomes are among the most highly diverse known?
A long but exciting journey that started with the decision not to work on the typical lab bacteria, but something from the real outside world!
You can download the complete model to try it for yourself or adapt to your own needs. We would also be happy to advice anyone wishing to grow and image cells in agarose mini beads. Finally, we hope our work inspires some of your own ideas.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
From RNA Detection to Molecular Mechanisms
Publishing Model: Open Access
Deadline: May 05, 2026
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: May 31, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in