Tracing the spread of carbapenem-resistant Klebsiella pneumoniae – the fastest-growing antibiotic resistance threat in Europe
Published in Microbiology

Klebsiella pneumoniae is a Gram-negative bacterium belonging to the Enterobacteriaceae family and is commonly found in natural and man-made environments. It has the impressive ability to colonise diverse animal species, including ourselves, our pets and livestock. Indeed, in healthy humans, K. pneumoniae is frequently found on the skin, in the mouth, and in the respiratory and gastrointestinal tracts. Yet K. pneumoniae is also an opportunistic pathogen and can cause a wide range of infections in humans (and other animals). These include urinary tract, respiratory and bloodstream infections, which are typically treated using beta-lactams and other antibiotics that are active against Enterobacteriaceae.
Of serious public health concern is that an increasing number of K. pneumoniae causing infections have become resistant to carbapenems, a class of beta-lactam antibiotics that is normally reserved as a last-line treatment option. A recent study reported that carbapenem-resistant K. pneumoniae represent the fastest growing antibiotic resistance threat in Europe, with a 6.16x increase in the number of attributable deaths between 2007 and 2015 (Cassini et al. 2019). Numerous other reports from around the world including from the US, China and Argentina (CDC, 2013; Gu et al. 2018; Gomez et al. 2011) have also highlighted the rapid emergence of carbapenem-resistant K. pneumoniae as a global problem. Together, these have led to the classification of carbapenem-resistant Enterobacteriaceae by the World Health Organisation as “critical” priority pathogens for which new antibiotics are urgently needed (WHO, 2017). Carbapenem-resistant K. pneumoniae infections have previously been associated with health care facilities (Falagas et al. 2007; Schwaber et al. 2011), implicating hospital transmission as a primary route of spread. However, recent studies have also highlighted a potential role for transmission of K. pneumoniae between humans and the environment (including animals), the latter which may serve as a reservoir of carbapenem resistance (Bush et al. 2011; Murray et al. 2018).
In April 2012, the European Centre for Disease Prevention and Control (ECDC) published an open call for tender for a European survey on carbapenemase-producing bacteria. The aim was to develop a consistent sampling framework and thereon investigate the incidence and epidemiology of carbapenemase-producing bacteria across Europe. Hajo Grundmann, then a Professor of Infectious Disease Epidemiology at the University Medical Center Groningen (UMCG) in the Netherlands, together with his team, won the bid and was tasked with coordinating and implementing the survey in conjunction with the ECDC. A total of 455 hospitals were recruited from 36 countries. After months of planning and training, the survey took place over six months between November 2013 and April 2014. Each hospital was asked to collect their first ten carbapenem non-susceptible clinical isolates of K. pneumoniae or E. coli as well as ten susceptible same-species comparator isolates. The results showed that the prevalence of carbapenem-resistant Enterobacteriaceae differed substantially with the highest burden in Mediterranean and Balkan countries, and also that carbapenem-resistant K. pneumoniae outnumbered E. coli by 11:1 (Grundmann et al. 2017).
The survey, known as EuSCAPE (European Survey of Carbapenemase-Producing Enterobacteriaceae), resulted in a remarkable collection of isolates representing the first unbiased snapshot of the European population of clinical K. pneumoniae and E. coli isolates from hospitalised patients. The isolate collection was subsequently whole genome sequenced, offering a wealth of information on the disease dynamics. At this point, I entered the project as a new Postdoctoral Scientist in the Centre for Genomic Pathogen Surveillance (CGPS) in Cambridge, UK, led by Prof. David Aanensen. Together with our international team of collaborators, we spent numerous hours (albeit very enjoyable!) during meetings in Freiburg, Florence, Cambridge and Bath, pouring over the data, which had endless avenues for exploration. In our study in Nature Microbiology, we focus on comparing the population structure of >1700 carbapenem-resistant and -susceptible K. pneumoniae isolates with relation to their geographic sampling locations. This has allowed us to draw inferences regarding their routes of spread that could have a meaningful impact on public health policy.
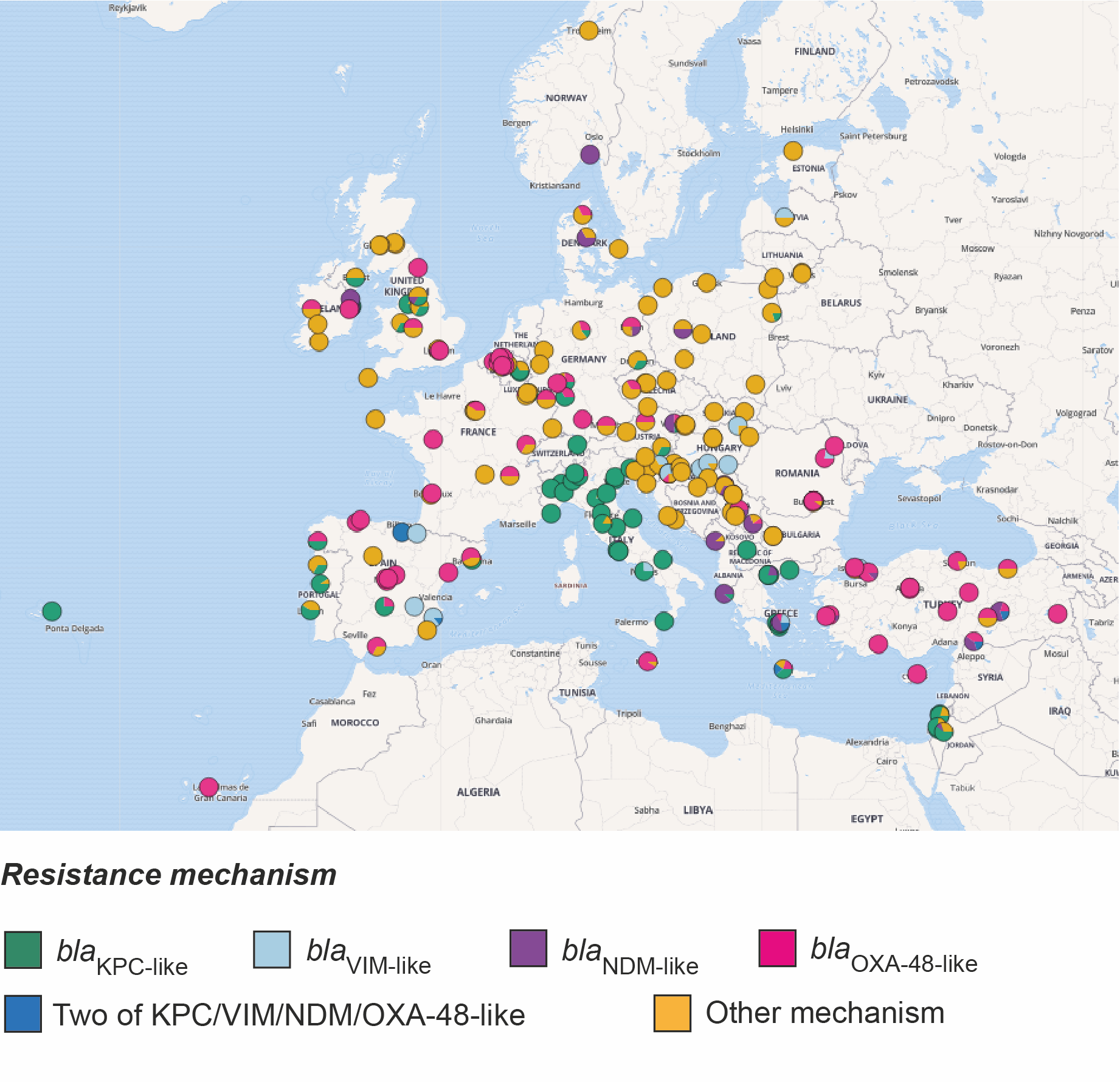
Firstly, since the two original categories of isolates, carbapenem-resistant and carbapenem-susceptible, mask differences in their resistance to other beta-lactam antibiotics, we divided the collection into five new categories. These are based on their genetic determinants of resistance to all beta-lactams and roughly reflect a decreasing scale of beta-lactam resistance. The most resistant category comprised isolates that possess one or more carbapenemase genes (blaKPC-like, blaNDM-like, blaVIM-like, blaOXA-48-like and blaIMP-like), the expression of which usually causes resistance to carbapenems. We noted that the majority of carbapenemase-positive isolates were confined to a small number of “high-risk” clones, which have emerged recently and spread rapidly. Furthermore, just over half (52.6%) of the carbapenemase-positive isolates were most closely related to another isolate in the collection obtained from a different patient in the same hospital. Given that the survey restricted collection to only 20 isolates per hospital (and thus not all patient isolates would have been included), these findings suggest frequent transmission between hospital patients. We then found that this percentage declined incrementally across the other four groups with decreasing levels of beta-lactam resistance. This suggests that the ecological constraints within the hospital favour spread of more resistant bacteria, likely a result of the empirical antibiotic treatments used.
We also found that carbapenemase-positive isolates were much more likely to be closely related to other isolates from a different hospital in the same country, rather than in a different country and that the genetic distance correlates with geographic distance between hospitals. These findings point to spread through national healthcare systems via referral networks, rather than spread via the environment or animals, the latter of which is less likely to respect hospital proximity and country borders. As previously, we found that the levels of between-hospital/within-country transmission also declined amongst the other four resistance groups. These findings fit with previous studies showing that K. pneumoniae infections with no or low antibiotic resistance are more likely to be community-acquired (Kang et al. 2006), and possibly caused by a strain already carried by the patient as part of the community-acquired microbiota.
Taken altogether, our findings indicate that the major driver of the carbapenem resistant K. pneumoniae epidemic in Europe has been transmission of hospital-adapted clones through healthcare networks. As a result, public health efforts should focus on enforcing effective infection control, including considering how patients move within and between hospitals. Identifying those hospitals that are central to regional or national referral networks, i.e. hospitals where many patients meet or their treatment pathways are crossing, would allow for better targeted control efforts. Thus, ramping-up surveillance and interventions at these selected sites holds the promise to turn sources into sinks and improve the resilience of entire health care systems against the further spread of antibiotic resistant bacteria.
References
- Cassini, A. et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: a population-level modelling analysis. Lancet Infect. Dis. 19, 56–66 (2019).
- Antibiotic resistance threats in the United States (CDC, 2013).
- Gu, D. et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect. Dis. 18, 37-46 (2018).
- Gomez, S. A. et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clinical Microbiology & Infection. 17, 1520-4 (2011).
- Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics (World Health Organization, 2017).
- Falagas, M. E. et al. Risk factors of carbapenem-resistant Klebsiella pneumoniae infections: a matched case-control study. J. Antimicrob. Chemother. 60, 1124–1130 (2007).
- Schwaber, M. J. et al. Containment of a country-wide outbreak of carbapenem-resistant Klebsiella pneumoniae in Israeli hospitals via a nationally implemented intervention. Clin. Infect. Dis. 52, 848–855 (2011).
- Bush, K. et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896 (2011).
- Murray, A. K. et al. Novel insights into selection for antibiotic resistance in complex microbial communities. mBio 9, e00969-18 (2018).
- Grundmann, H. et al. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect. Dis. 17, 153–163 (2017).
- Kang, C-I. et al. Community-acquired versus nosocomial Klebsiella pneumoniae bacteremia: Clinical features, treatment, outcomes, and clinical implication of antimicrobial resistance. Journal of Korean Medical Science. 21, 816-822 (2006).
Follow the Topic
-
Nature Microbiology

An online-only monthly journal interested in all aspects of microorganisms, be it their evolution, physiology and cell biology; their interactions with each other, with a host or with an environment; or their societal significance.
Related Collections
With Collections, you can get published faster and increase your visibility.
The Clinical Microbiome
Publishing Model: Hybrid
Deadline: Mar 11, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in