Tracking mobile elements between commensals and pathogens in the gut microbiome
Published in Microbiology

Antibiotic resistance genes often reside on mobile genetic elements (MGE) that are transferred between organisms, including pathogens, presumably within the gut microbiome. Plasmids, in particular, are frequently implicated in conferring multi-drug resistance to pathogens. Yet, understanding which species carry plasmids carrying antibiotic resistance within a microbiome has been difficult because these associations are lost during both short- and long-sequencing and cannot be recovered by genome assembly alone.
To resolve bacteria-MGE associations, we applied untargeted proximity ligation sequencing (Hi-C) (see below) to gut microbiome samples, creating physical links between chromosomal DNA and MGEs that can be observed by sequencing. We applied this approach to identify multi-drug resistant plasmids within a cohort of patients with neutropenia. With compromised immune systems, these patients are uniquely susceptible to multi-drug resistant infections, and samples collected from these patients during the course of their hospitalizations allowed us to ask whether horizontal transfer of mobile elements could be observed in each person through time.
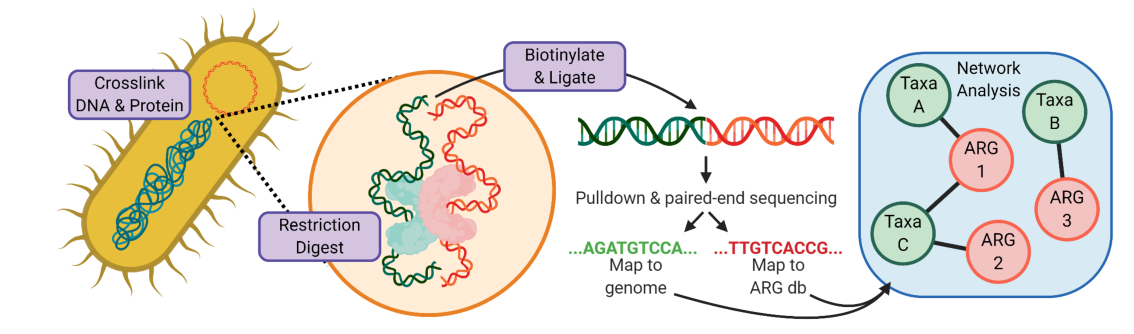
As a test of this method, which has only recently been applied to bacteria and microbiomes, we performed a rarefaction analysis to determine whether we were sequencing our Hi-C amplicons to a suitable depth to exhaustively capture mobile gene-host associations.
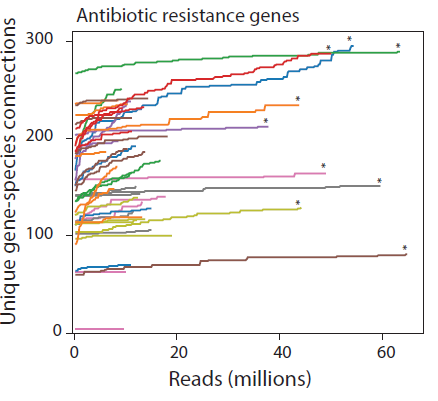
Working under the assumption that most transfer events involve MGEs that are endogenous to the microbiome, we sequenced the first time point of each time-course (starred above) extremely deeply. Therefore, any new associations identified during the timecourse between genes and organisms that existed at the start, might indicate newly horizontally transferred genes. We knew this was a provocative claim, so we set super-strict criteria: both the donor and recipient organisms had to be assembled during the first timepoint into high quality metagenome-assembled genomes (MAGs); the mobile gene must clearly associate with the donor at the start of the time course (mean = 28.6 Hi-C reads) and not at all (zero reads) with the recipient, nor with any recipient genome within a concordant higher taxa; and then must clearly associate with the recipient taxa in the latter timepoints (mean = 14.8 Hi-C reads). We then looked for cases where we saw the same recipient associations across multiple latter timepoints (19 out of 155 showed this) and for those which could be observed across multiple contigs (roughly 1/3), providing further support of these observations.
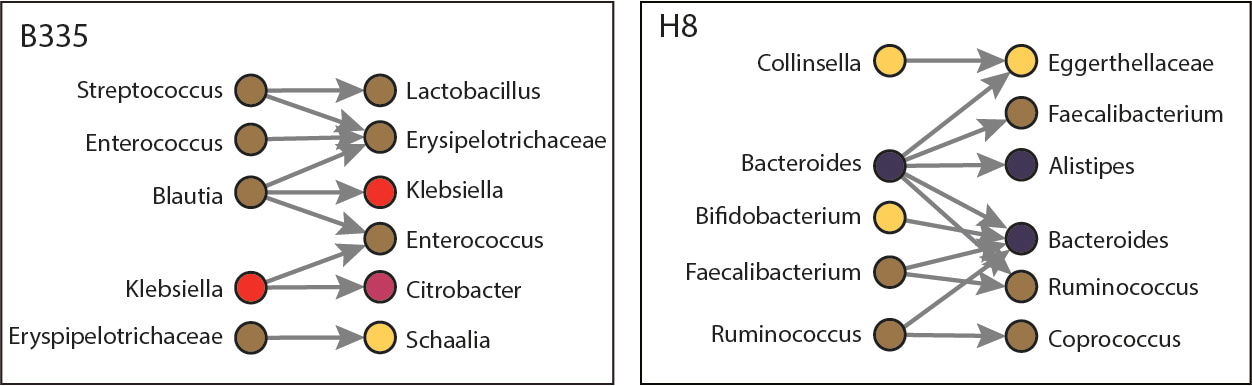
Incredibly, we witnessed pathogens donating antibiotic resistance genes to members of the human microbiome, expanding the reservoir of resistance genes. We also observed pathogen acquisition of antibiotic resistance genes that have been associated with multi-drug resistance in clinically settings. We saw this not only in neutropenic patients (like B335 below) but also in human individuals (H8 below), albeit transfers in healthy patients occurred between commensals. Our work reveals a network of horizontal gene exchange that is perpetual within our microbiomes, with potentially damning consequences for the spread of multi-drug resistant plasmids.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in