Two Omicron sublineages, one patient: the shortest case of reinfection so far
Published in Healthcare & Nursing, Microbiology, and Protocols & Methods
At times, scientific writing is also about telling interesting stories. Obviously, these need to be relevant in the scientific context, being valuable for discussion for other studies. In the midst of facing a pandemic due to an emerging virus, there are stories that can serve to generate insights even on control measures. This was the situation that led us to study and report this case of such a rapid reinfection with the BA.1.1 and BA.2 strains in a patient in southern Brazil.
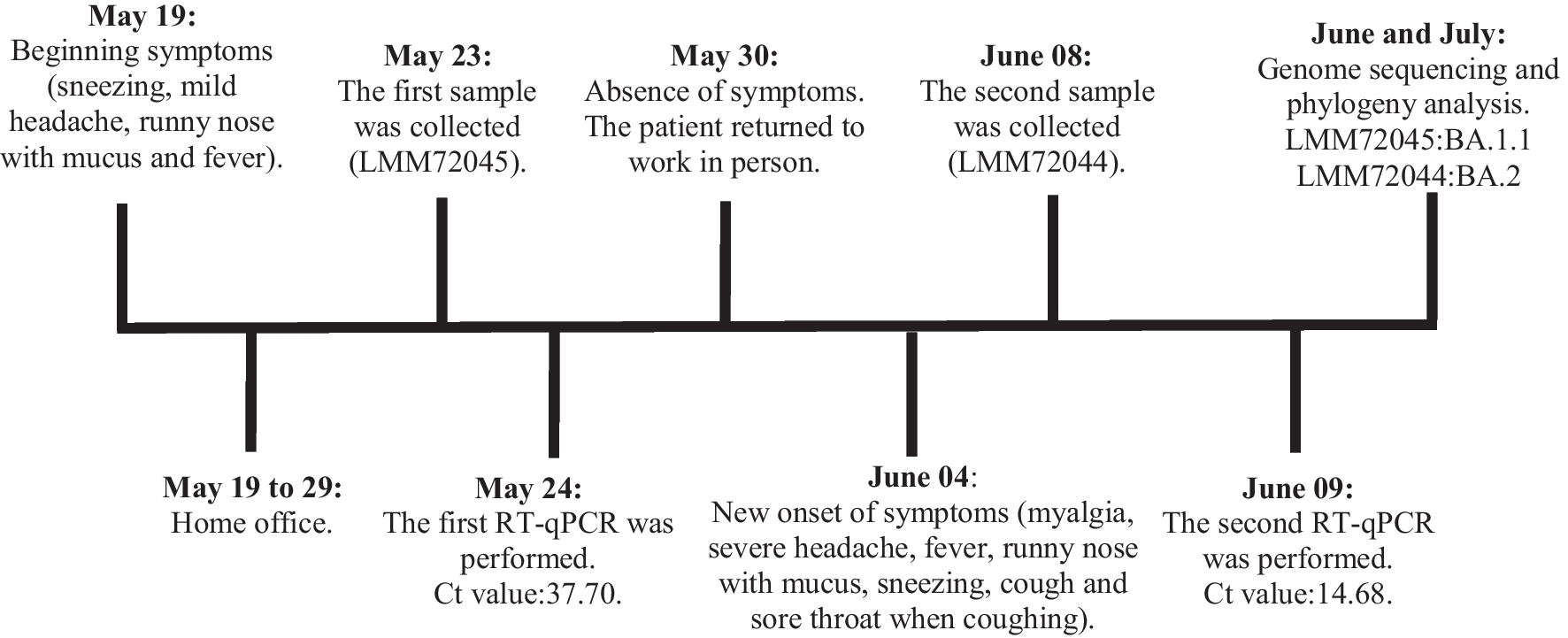
The patient in this clinical case experienced symptoms of COVID-19 twice, with the first round of symptoms starting on May 23rd 2022 and the second round starting on June 4th (Fig. 1). The 52-old man, triple vaccinated, had clinically recovered completely, tested negative in RT-qPCR tests after a few days of mild symptoms. He has happily returned to work in office and then began to present more intense symptoms a few days later. The viral detection assay was positive again. Both the health professionals and surveillance teams involved, as we in the laboratory, obviously thought of a prolonged infection with a relapse.
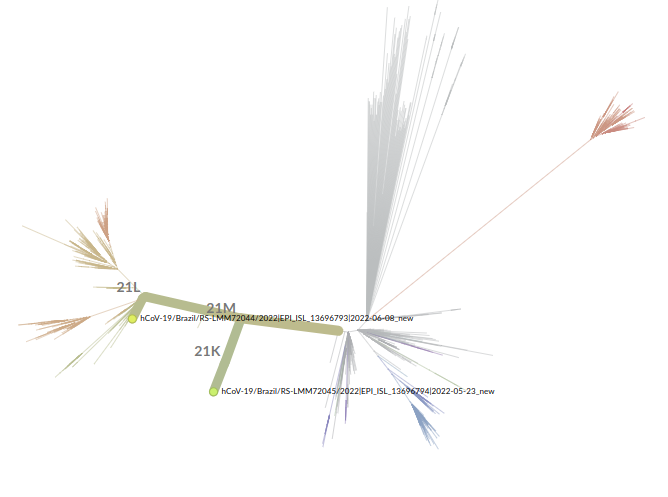
Since we had the SARS-CoV-2 positive samples collected on those two occasions, we thought it would be quite interesting to perform genome sequencing on both to check whether the virus genome had suffered any mutations over that period (although the negative assays in the middle of monitoring were still a mystery at that point). Surprisingly, the results showed that the patient was infected with two different sublineages of the Omicron variant of SARS-CoV-2: BA.1.1, for the first round of symptoms, and BA.2 for the second (Table 1).
Table 1. Sequencing results (reads, coverage, and length) and signature mutation at spike protein for both samples evaluated
|
|
|
Samples |
|
|---|---|---|---|
|
|
|
LMM720451 |
LMM720442 |
Reads |
Total reads |
242,155 |
326,802 |
Coverage |
85.3% |
99.5% |
|
Length (mean) |
130 bp |
133 bp |
|
Signature mutation at spike protein3 |
Total |
36 |
31 |
Shared mutations4 |
13 |
21 |
|
Specific mutations |
15 |
10 |
|
Mutations detected |
19 |
31 |
|
Fortunately, the patient recovered. What we learned and wanted to tell other researchers is that people's individual responses to an infection can indeed vary, and that the Omicron variant and its variants can surprise us in terms of their ability to generate reinfections. Also, it's not by chance that we probably see so many SARS-CoV-2 recombinants out there nowadays since co-infections must also be very likely. A lot of good sense and cautiousness is needed when you have a large number of cases of COVID-19 going on in your community (the same must be true for other respiratory viral infections), and general measures of respiratory hygiene must be adopted even by patients that have recently recovered.







Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in