UBS-seq: An ultrafast bisulfite sequencing method more accurately detecting 5-methylcytosine in DNA and RNA
Published in Bioengineering & Biotechnology
5-methylcytosine (5mC) in DNA is a fundamental epigenetic mark that controls gene expression in numerous biological processes.[1] In human, the abnormal methylation patterns of 5mC reflect disease status and provide effective biomarkers for early diagnosis and monitoring of human diseases. Accurate detection of 5mC sites is critical for both basic research and clinical applications.[2] Due to its high resolution, robust reproducibility and low cost, bisulfite sequencing (BS-seq) technology has been the gold standard for DNA 5mC detection, and has been widely used in basic research and clinic practices.[3] However, the current BS-seq method for DNA 5mC detection still suffers from several notable limitations: (1) long reaction time, the widely adopted conventional BS treatment requires ~3 hours, limiting its application in rapid clinic diagnosis; (2) in high GC DNA regions or highly structured DNA (such as mitochondrial DNA), C-to-U conversion could be incomplete, resulting in high background and false positives; (3) severe DNA damage, limiting its application for low input samples such as circulating cell-free DNA (cfDNA); (4) BS treatment leads to more severe degradation of non-methylated regions, resulting in overestimation of methylation levels. A rapid and accurate detection of 5mC as a disease marker using low input samples in clinical practice would be ideal. Additionally, BS-seq for RNA m5C detection also faces many challenges due to instability of RNA and presence of relatively stable secondary structures in different RNA regions, which limits research on functional investigations of RNA m5C in diverse cell types.
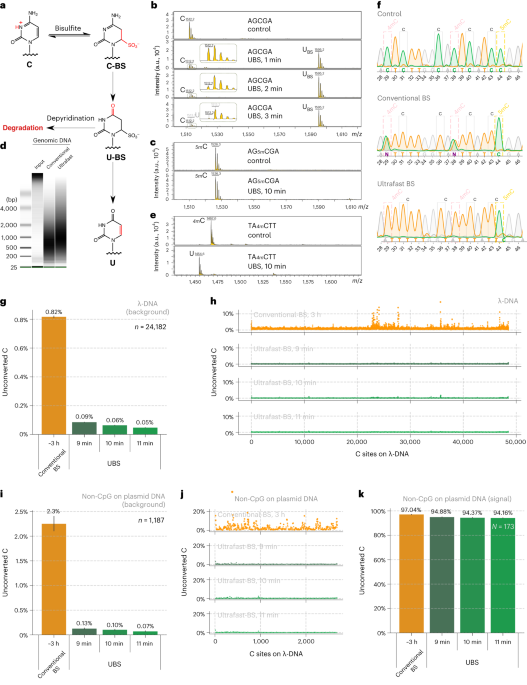
Based on the mechanism of BS-seq and the DNA degradation mechanism caused by the BS reaction, we found that a new BS recipe composed of ammonium instead of sodium salts can greatly improve the BS efficiency so that complete C-to-U conversion can be achieved rapidly while 5mC remains unchanged (Fig. 1a), with DNA degradation significantly reduced (Fig. 1b). Sequencing unmodified lambda DNA revealed that the background noise could be reduced by ~14 times compared to conventional BS-seq and the overall conversion efficiency of UBS-seq is also more consistent (Fig. 1d).
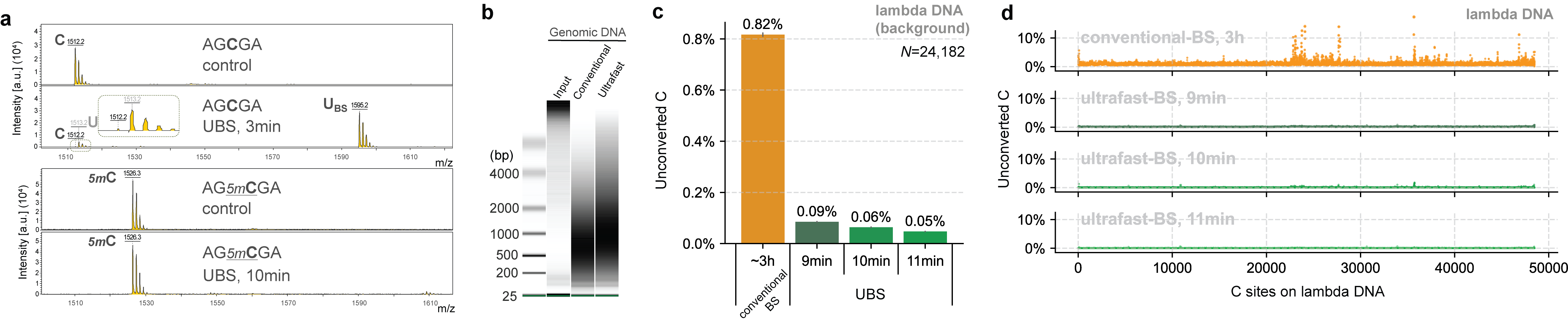
Fig. 1. UBS-seq is highly effective in DNA 5mC detection. (a) Maldi TOF MS showed that complete C-to-U conversion could be achieved within 3 minutes when a C-5mer DNA model oligo was treated with UBS recipe at 98 ºC while the corresponding 5mC oligo remained unchanged. (b) UBS treatment caused less DNA damage than the conventional BS treatment. (c) UBS-seq afforded much lower unconverted C ratio than conventional BS-seq. (d) UBS-seq resulted in evenly distributed low background across the tested genome while conventional BS-seq caused uneven high backgrounds at certain regions.
UBS-seq is effective on sequencing low input samples while reducing background and 5mC level overestimation. It can also be directly used without prior extracting gDNA using 1 to100 mESC cells. We also applied UBS-seq to cfDNA samples extracted from the blood of early-stage colorectal cancer patients and controls, and found differentially methylated regions as potential biomarkers for colorectal cancer early diagnosis. These results suggest that UBS-seq has broad application potentials in 5mC biomarker discovery and application, in particular from limited input samples. It could also be used for rapid clinical diagnosis and real-time decision-making in surgery.
UBS-seq can also be used to more accurately detect m5C in RNA. m5C exists in various RNA species, affecting a variety of cell functions and playing critical roles in various human diseases. Because RNA is much easier to be degraded under BS conditions and the presence of highly stable secondary structure in certain RNA regions, conventional BS treatment has difficulties in detecting RNA m5C in low abundant or highly structured RNA species. Compared with 5mC in mammalian gDNA, both the modification sites and modification levels of m5C in mRNA are much lower. Therefore, the main challenge of RNA BS-seq is how to accurately detect and quantify m5C sites while avoiding false positives.[4] We found a slightly altered UBS-seq recipe that can reduce the background noise to < 1% (Fig. 2a), which is much lower than those reported from several previously published m5C BS-seq protocols (Fig. 2b). When applying UBS-seq to stable and highly structured tRNAs, we detected accurate m5C stoichiometry at several known m5C sites (Fig. 2c). When employing UBS-seq to sequence mRNA isolated from HeLa and HEK293T cells, we detected more than 2,000 m5C sites with modification ratios of >5%, with many sites sharing conserved motifs reported previously (Fig. 2d). Since UBS-seq has very low background noise, we also detected a large number of sites with low modification stoichiometry, and their stoichiometry responded to NSUN2 knockout (Fig. 2e) and could be rescued by transfecting NSUN2 gene back to the knockout cells, further validating these m5C sites (Fig. 2f). These results demonstrated that the m5C UBS-seq method is not only very sensitive and robust, but also accurate.
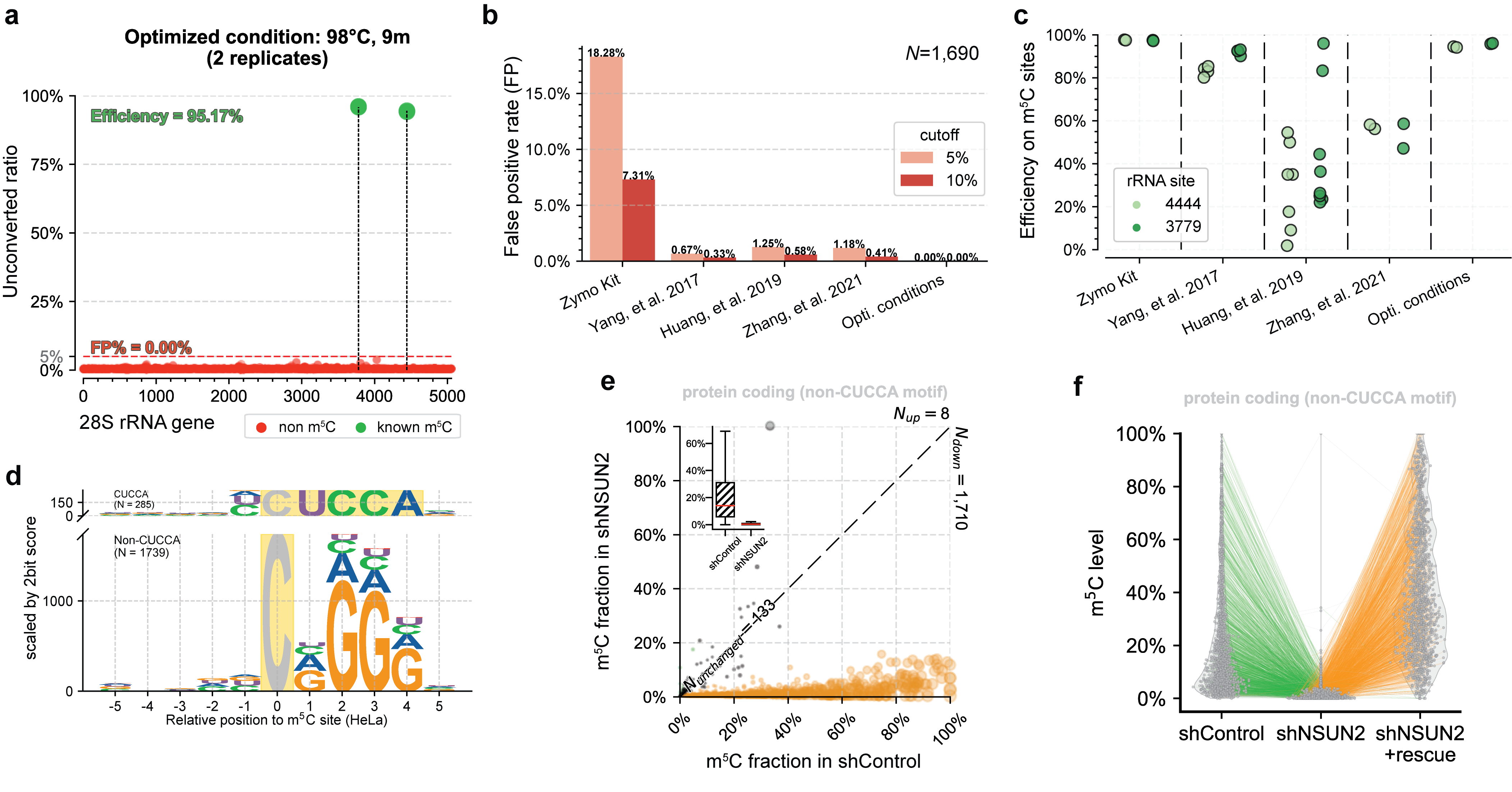
Fig. 2. UBS-seq accurately detects m5C in RNA. (a) UBS-seq detected the two known m5C sites with high signal-to-noise. (b) Comparison of the false positives between UBS-seq and other methods. (c) Comparison of the m5C detection between UBS-seq and other BS protocols. (d) UBS-seq detected the conserved motifs for NSUN2 and NSUN6 substrates. (e) ~90% m5C sites detected in HeLa mRNA responded to NSUN2 KO. (f) Majority of m5C site signals were rescued when NSUN2 gene was introduced back to NSUN2 KO cells.
[1] Dor, Y. & Cedar, H. Principles of DNA methylation and their implications for biology and medicine. The Lancet 392, 777–786 (2018).
[2] Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023).
[3] Frommer, M. et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proceedings of the National Academy of Sciences 89, 1827–1831 (1992).
[4]Squires, J. E. et al. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 40, 5023–5033 (2012).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in