Discovery of a Unified Autism Circuit Signature via AI-Powered Whole-Brain Analysis
Published in Neuroscience, Computational Sciences, and General & Internal Medicine
Autism spectrum disorder (ASD) is a neurodevelopmental condition primarily characterized by deficits in social communication, as well as restricted and repetitive patterns of behavior and sensory processing abnormalities. In addition to these core features, individuals with ASD often present with a range of comorbidities. Because symptoms can vary widely, ASD is described as a “spectrum.” A mix of genetic factors, biological sex, and environmental conditions influences these differences. Understanding the brain mechanisms behind ASD is key to developing better ways to diagnose and treat it.
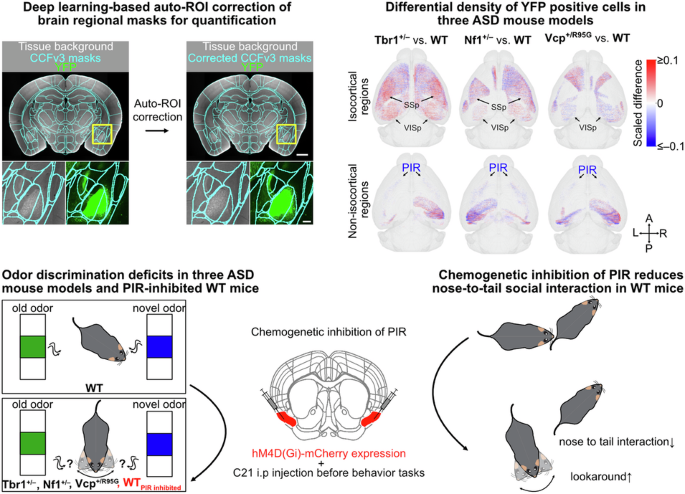
One key biological process implicated in ASD is altered brain connectivity. However, because different ASD-associated mutations may lead to distinct patterns of circuit rewiring, identifying common circuit mechanisms underlying the core symptoms remains a major challenge. To address this, we developed BM-auto (Brain Mapping with auto-ROI correction), an AI-powered whole-brain mapping and quantification platform optimized for fluorescence imaging of serial brain slices, to efficiently process the whole-brain data and collect accurate results for analysis. It greatly accelerates our progress in whole-brain connectivity analysis in mouse models.
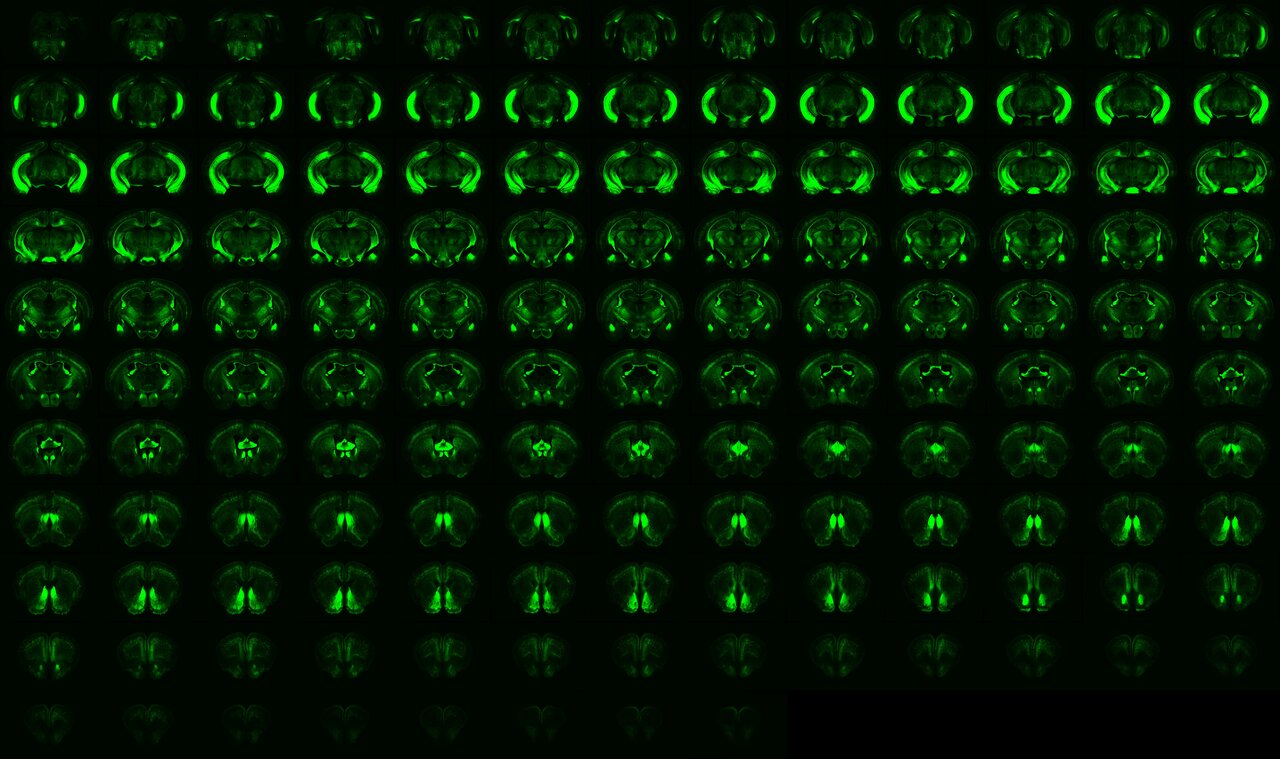
Using standard immunohistochemistry protocols for thin cryosections, our workflow was optimized for high-throughput processing of hundreds of slices. Brain slices were imaged using a high-content system, registered to the Allen Common Coordinate Framework version 3 (CCFv3), and segmented for quantification. To improve the accuracy of regional quantification, we first manually adjusted the regional masks in each hemisphere showing variations in YFP signals or YFP cell distributions across all samples.
This manual correction of regional masks was both time-consuming and labor-intensive: correcting 37 brain regions across more than 72 hemispheres required nearly three months of effort, and over 400 brain regions remained unvalidated. Fortunately, we collaborated with Dr. Chien-Yao Wang, an AI expert and Associate Research Fellow at the Institute of Information Science, Academia Sinica. Using our manually corrected masks and corresponding background images, Dr. Wang’s team trained a deep learning model capable of automatically adjusting the original CCFv3 regional masks to match true anatomical boundaries. This approach now enables highly reliable, automated region-wise quantification across the entire dataset.
With this powerful tool, we investigated brain connectivity changes across three ASD-related monogenic mouse models, including Tbr1+/–, Nf1+/–, Vcp+/R95G mice. To broadly examine connectivity changes beyond specific circuits, we crossed each mutant line and its wild-type littermates with Thy1-YFP reporter mice. By examining brain-wide YFP expression in neuronal soma, neuropil, and long-range axons of a subset of neurons, such as cortical deep-layer projection neurons, hippocampal neurons, amygdala neurons, and some midbrain neurons, we mapped the shared circuit mechanisms linking to ASD.
When we compared YFP signal patterns and the number of YFP⁺ cells between mutant and wild-type mice, we found that Tbr1, Nf1, and Vcp deficiencies each affected YFP signals and cell distributions in different ways. Notably, cortical regions involved in sensory processing were consistently affected across all three ASD models. However, the specific hemispheric regions impacted, as well as the direction and extent of these changes, varied among the three mutations. To understand how these changes might reflect altered brain circuits, we examined how Thy1-YFP circuits—which connect projecting and target regions—might be rewired in each ASD-related mutation.
We first identified brain regions where differential YFP signals primarily resulted from altered YFP⁺ axonal innervations. These regions were predominantly located in the thalamus, hypothalamus, midbrain, and hindbrain. To determine the candidate upstream regions contributing to these changes, we integrated our data with the spatial search function of the Allen Mouse Brain Connectivity Atlas. For each region showing differential YFP axon targeting, we retrieved the repertoire of potential upstream regions along with their relative axonal projection densities at the sites of maximum YFP difference. We then filtered these candidate upstream regions to include only those that also exhibited abnormal YFP⁺ cell distributions in ASD mutant mice.
Using this information, we built predicted Thy1-YFP circuit maps for the following comparisons: Tbr1+/– vs. Tbr1+/+, Nf1+/– vs. Nf1+/+, and Vcp+/R95G vs. Vcp+/+. The results showed distinct patterns of circuit rewiring. Tbr1 deficiency tended to increase innervation by Thy1-YFP projection neurons. Nf1 deficiency had mixed effects, with some pathways strengthened and others weakened. Vcp+/R95G mice generally showed reduced Thy1-YFP innervation. We confirmed some of these predicted changes in Tbr1+/– mice. In particular, we observed stronger connections from the somatosensory cortex to the ventral posterior thalamus and from the primary visual cortex to the dorsal lateral geniculate nucleus, as shown by CTB retrograde tracing.
Despite the distinct Thy1-YFP circuit rewirings observed across the three ASD mutations, we identified a convergent deficit shared among them. The piriform area (PIR)— a brain region involved in processing smells — consistently exhibited impairments, characterized by reduced YFP signals and a decreased number of YFP⁺ neurons. This convergent PIR deficit aligns with the behavioral abnormalities that display deficits in odor discrimination while maintaining normal olfactory sensation.
We also examined whole-brain neuronal activity by measuring C-FOS expression in Vcp+/R95G mice after odor stimulation. Similar to our previous findings on impaired PIR neuronal activity in Tbr1+/– mice, PIR neuronal responses were also impaired in Vcp+/R95G mice following odor stimulation. In addition, overall neuronal activity across the brain was reduced in response to odor, further supporting the idea of predicted defective whole-brain neural connectivity in Vcp+/R95G mice. To probe causality between PIR activity and behavior phenotype, we suppressed bilateral PIR activity in wild-type mice by a chemogenetic approach. Inhibition led to deficits in odor discrimination, reduced odor sensitivity, and decreased anogenital sniffing during social interactions, demonstrating that PIR activity is critical for odor processing and specific social behaviors.
In summary, this study has two achievements. First, we established an efficient and accurate BM-auto pipeline for mesoscopic whole-brain imaging data, which can be widely applied to examine diverse molecules as long as they can be visualized using different methods on brain sections. Second, we identified convergent PIR deficits across various ASD models using the BM-auto pipeline. While different mutations lead to unique patterns of circuit rewiring, PIR impairments consistently appear as a shared neural feature associated with sensory and social behavioral challenges in ASD. Further research is necessary to explore how PIR dysfunction influences ASD traits during development, helping to clarify how sensory deficits evolve into core symptoms of ASD.
Follow the Topic
-
Molecular Psychiatry

This journal publishes work aimed at elucidating biological mechanisms underlying psychiatric disorders and their treatment, with emphasis on studies at the interface of pre-clinical and clinical research.




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in