Understanding the Formation of Natural Product Toxins in Pathogenic Bacteria
Published in Chemistry

Burkholderia mallei and Burkholderia pseudomallei, which comprise the B. pseudomallei group of bacteria, are notorious human pathogens.1 Both bacteria show high mortality rates in human infections and are highly recalcitrant to antibiotic treatment.2 This has prompted extensive research into the molecular mechanisms of infection with the goal of disarming the pathogens on a molecular scale by antivirulence therapy.2 The work published in Nature Chemistry by the Groll and Hertweck groups reports on a cyclopropanol synthase (BurG) that builds the warhead of the malleicyprols, which are small molecule toxins crucial to the virulence of B. pseudomallei group bacteria. In-depth knowledge on the formation of the key cyclopropanol ring potentially lays the foundation for the development of anti-virulence agents to combat these pathogens.
Burkholderic acid originates from the virulence-associated biosynthetic gene cluster bur
The first steps towards elucidating a novel enzymatic route to cyclopropanol formation came with our discovery of the small molecule burkholderic acid,3 a finding reported simultaneously by the group of Sean F. Brady.4 The identification of burkholderic acid (syn. malleilactone, as named by the Brady group) was the result of the efforts of both groups to awaken silent biosynthetic gene clusters (BGCs) from B. thailandensis. The natural product repertoire of B. thailandensis piqued our interest due to its close relation to B. mallei and B. pseudomallei.5 The Brady laboratory designated the bur BGC (common to the genomes of B. mallei, B. pseudomallei and B. thailandensis) as a virulence determinant by testing a bur knockout strain of B. thailandensis in eukaryotic infection models based on the nematode Caenorhabditits elegans and the amoeba Dictyostelium discoideum. Yet surprisingly, burkholderic acid, the main isolated product of the bur BGC, is not cytotoxic, meaning that the manner in which the bur BGC contributes to virulence remained unexplained (Fig. 1).
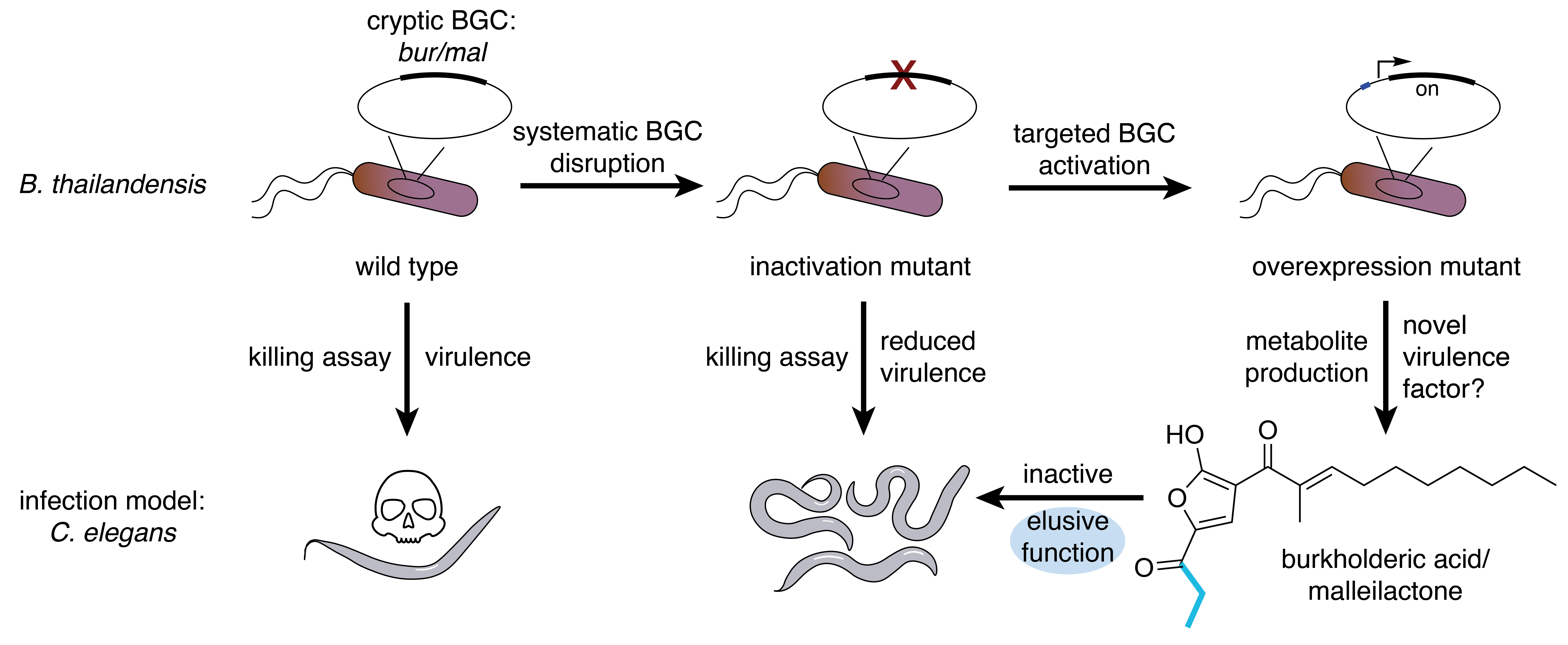 Fig. 1. Identification of the bur biosynthetic gene cluster (BGC) as a virulence determinant in B. thailandensis, a model organism for the human pathogens B. mallei and B. pseudomallei. Targeted activation of the bur cluster led to the identification of burkholderic acid. Although a bur knockout strain of B. thailandensis was avirulent, purified burkholderic acid was not cytotoxic. The propanone chain of burkholderic acid, which is derived from methionine, is coloured blue.
Fig. 1. Identification of the bur biosynthetic gene cluster (BGC) as a virulence determinant in B. thailandensis, a model organism for the human pathogens B. mallei and B. pseudomallei. Targeted activation of the bur cluster led to the identification of burkholderic acid. Although a bur knockout strain of B. thailandensis was avirulent, purified burkholderic acid was not cytotoxic. The propanone chain of burkholderic acid, which is derived from methionine, is coloured blue.
A closer inspection of the bur BGC revealed a complex genomic architecture with a vast excess of encoded biosynthetic enzymes than seemingly necessary for the formation of burkholderic acid. This raised the possibility that further natural product diversity originating from the BGC could be behind the virulent phenotype of B. thailandensis. We were also intrigued by an apparent biosynthetic illogicality; the simple C3 propanone chain of burkholderic acid arises from truncation of methionine rather than the energetically less-demanding use of propionate as a building block.
The true virulence factor produced by the bur/mal assembly line possesses a cyclopropanol warhead
We set out to determine the enigmatic role of the bur BGC in the virulence of B. pseudomallei group bacteria. Reexamination of the metabolic profile of B. thailandensis revealed a suite of compounds, named malleicyprols, that showed high structural resemblance to burkholderic acid.6 These compounds differed from burkholderic acid in one striking detail – the presence of a cyclopropanol moiety instead of the puzzling propanone unit. Subsequent cytotoxicity assays established the malleicyprols as the true virulence factors of the bur BGC (Fig. 2). Our finding that degradation of the malleicyprols leads to the formation of burkholderic acid offered a plausible explanation for the counter-intuitive use of methionine; methionine is the biosynthetic precursor of the cyclopropanol unit of the malleicyprols, while its incorporation into the propanone unit of burkholderic acid can be rationalised by malleicyprol degradation.
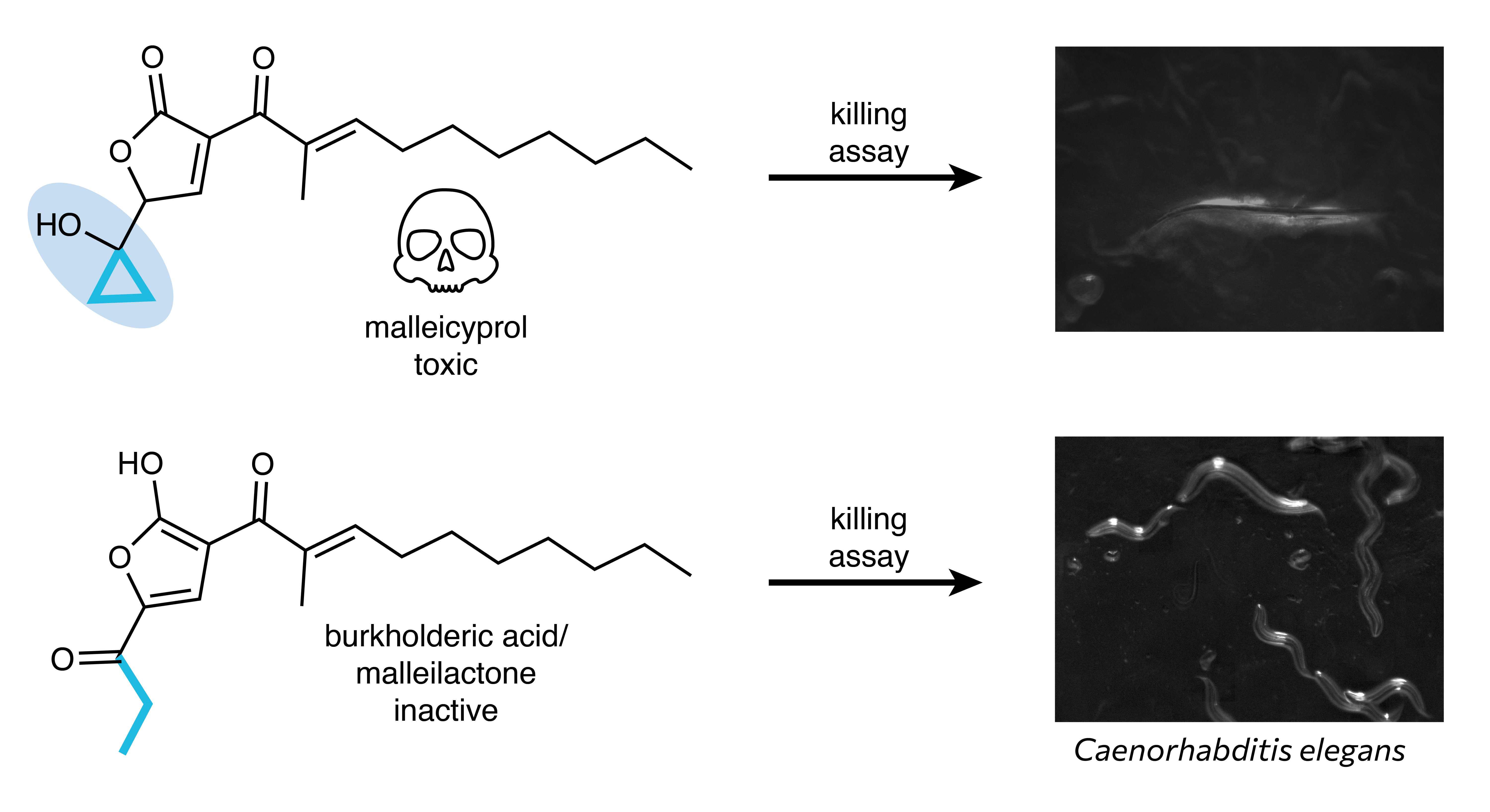
Fig. 2. The malleicyprols (a representative malleicyprol from the suite is shown) possess a cyclopropanol warhead. The cyclopropanol-substituted malleicyprols are active against C. elegans while burkholderic acid is inactive. The propanone chain of burkholderic acid and the cyclopropanol unit of the malleicyprols, which are derived from methionine, are coloured blue.
Since the bioactivity of the malleicyprols could be pinpointed to the unusual cyclopropanol unit, we reasoned that specific inhibition of the formation of this cyclopropanol warhead from methionine could provide a unique opportunity for targeted suppression of B. pseudomallei and B. mallei virulence. To form the basis for research in this direction, we sought to elucidate the intermediates and enzymatic mechanisms involved in cyclopropanol formation.
Sulfonium acids as key intermediates in the formation of the cyclopropanol warhead
Through careful mass spectrometric examination of extracts from a library of bur gene deletion mutants, we identified various sulfonium acids as biosynthetic intermediates that accumulated in specific mutants.7 Notably, the sulfonium group is a well-known structural motif employed in organic synthesis to construct strained three-membered rings by using dimethyl sulfide (DMS) as a leaving group. In the biosynthetic pathway to the cyclopropanol moiety of the malleicyprols, DMS already becomes evident as a leaving group in the first step, where methionine is converted to the sulfonium acid S-methylmethionine (SMM). Much of the complexity of the bur BGC is accounted for by the various additional steps including truncation, elongation and hydroxylation of the SMM precursor, that need to occur before the cyclopropanol ring is finally formed from gonydiol (Fig. 3).
Fig. 3. Sulfonium acid intermediates and enzymes involved in the biosynthetic pathway to the cyclopropanol moiety of the malleicyprols explain the complex architecture of the bur BGC. BurB: methyltransferase, BurI: decarboxylase, BurD: aminotransferase, BurE: dehydrogenase, BurA: NRPS-PKS hybrid, BurC: hydroxylase, SMM: S-methylmethionine, DMSP: dimethylsulfoniopropionate.
Mechanism of cyclopropanol formation
The hydroxylation reaction that installs the α-hydroxy moiety of gonydiol (Fig. 3) generates the focal point for cyclopropanol formation. As detailed in the current study8, the cyclopropanol synthase BurG catalyses a transient oxidation at this α-position and thus generates a nucleophilic enediol. Intramolecular attack of this enediol leads to the loss of DMS and cyclopropanol formation. The resulting ketone is reduced by BurG, thereby generating the final cyclopropanol-containing building block trigonic acid and leaving the overall reaction redox neutral (Fig. 4). Protein crystallography offered unique snapshots of this central step of malleicyprol biosynthesis and revealed its cryptic redox chemistry.
Fig. 4. Simplified enzymatic mechanism of cyclopropanol formation through a cryptic redox cycle catalysed by the cyclopropanol synthase BurG. DMS: dimethyl sulfide.
Perhaps most importantly, the discovery of BurG as a key enzyme in the bur assembly line coupled with the elucidated 3D structures offer valuable knowledge for the development of selective inhibitors to combat B. pseudomallei group pathogens, for instance by rational design. Promisingly, the risk of resulting drug candidates having off-target effects is likely lessened by the fact that humans do not encode BurG homologues.
References
1. Wiersinga, W. J. et al. Melioidosis. Nat. Rev. Dis. Prim. 4, 17107, (2018).
2. Estes, D. M., Dow, S. W., Schweizer, H. P. & Torres, A. G. Present and future therapeutic strategies for melioidosis and glanders. Expert. Rev. Anti. Infect. Ther. 8, 325-338, (2010).
3. Franke, J., Ishida, K. & Hertweck, C. Genomics-driven discovery of burkholderic acid, a noncanonical, cryptic polyketide from human pathogenic Burkholderia species. Angew. Chem. Int. Ed. 51, 11611-11615, (2012).
4. Biggins, J. B., Ternei, M. A. & Brady, S. F. Malleilactone, a polyketide synthase-derived virulence factor encoded by the cryptic secondary metabolome of Burkholderia pseudomallei group pathogens. J. Am. Chem. Soc. 134, 13192-13195, (2012).
5. Biggins, J. B., Kang, H. S., Ternei, M. A., DeShazer, D. & Brady, S. F. The chemical arsenal of Burkholderia pseudomallei is essential for pathogenicity. J. Am. Chem. Soc. 136, 9484-9490, (2014).
6. Trottmann, F. et al. Cyclopropanol warhead in malleicyprol confers virulence of human- and animal-pathogenic Burkholderia species. Angew. Chem. Int. Ed. 58, 14129-14133, (2019).
7. Trottmann, F. et al. Sulfonium acids loaded onto an unusual thiotemplate assembly line construct the cyclopropanol warhead of a Burkholderia virulence factor. Angew. Chem. Int. Ed. 59, 13511-13515, (2020).
8. Trottmann, F. et al. Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead. Nat. Chem. https://doi.org/10.1038/s41557-022-01005-z (2022).
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in