Unravelling how the stress sensor ATF6 stirs up trouble (and tumors)
Published in Cancer, General & Internal Medicine, and Anatomy & Physiology
We previously generated a transgenic murine model with chronic activation of the endoplasmic reticulum unfolded protein response (UPRER) activating transcription factor 6 (ATF6) in the intestinal epithelium. Turns out, when ATF6 gets stuck in “on” mode, things get very interesting. Numerous studies recognize the UPRER as a fundamental mediator in cellular physiology and the pathogenesis of inflammatory disorders, metabolic diseases and cancer, including colorectal cancer (CRC). At the time, our transgenic murine model gave us two valuable and novel insights into the role of the ATF6 signaling pathway: 1. Chronic ATF6 activation in the intestinal epithelium leads to spontaneous (no carcinogens, no tinkering) tumor formation in the colon and (here’s the twist) 2. Tumorigenesis is microbiota dependent. The question that remained was, how does ATF6 turn intestinal cells into tumor factories? So, we set out on a 7-year scientific saga to understand the mechanisms involved.
From mice to humans: is ATF6 a real player in CRC?
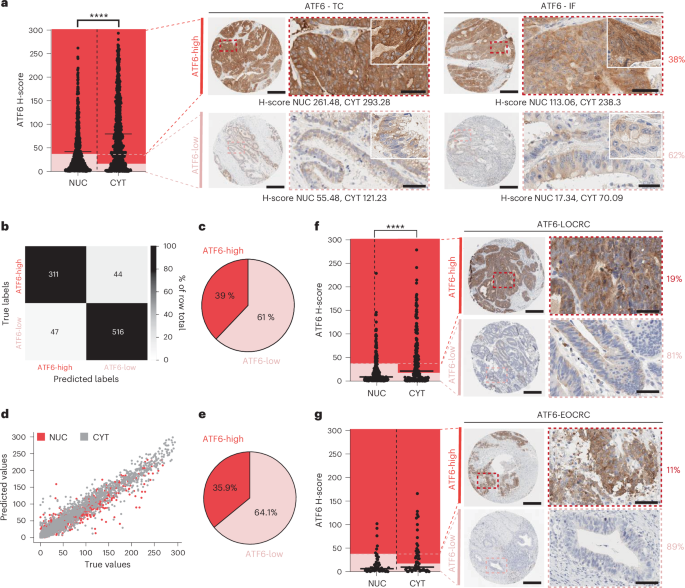
In a first step we wanted to elucidate the human relevance of ATF6 activation in the context of CRC. Through immunohistochemistry staining of independent German CRC patient cohorts, we could show that a subpopulation of up to 38% of CRC patients can be classed as expressing high levels of ATF6. That’s not a niche club, it’s a sizeable crowd. In a hypothesis-free approach, digging through whole exome sequencing and mRNA sequencing data of The Cancer Genome Atlas Database (TCGA), we associated high ATF6 expression to correlate with the presence of CRC-enriched bacterial genera. Conclusion: yes, ATF6 activation shows human relevance in CRC.
What is happening between chronic ATF6 activation and tumor formation?
To unravel the mystery, we went full systems biology: mRNA sequencing, untargeted metabolomics and 16S rRNA sequencing. Using intestinal organoid systems, germ-free mice, specific-pathogen-free mice as well as CRC patient biopsies we could confidently add two further insights to our list: 3. ATF6 alters colonic lipid metabolism in the presence of bacteria, 4. ATF6 drives fatty acid elongation, with long chain fatty acids accumulating in mice and in CRC patients. Furthermore, through extensive experiments in mice by first inhibiting fatty acid synthase and subsequently performing microbiota transfer experiments into germ-free mice using cecal content from these inhibitor-treated mice we showed that 5. ATF6-related lipid profiles are causally linked to tumor-inducing microbiota. Conclusion: ATF6 alters lipid profiles, and these lipids not just metabolic wallpaper but associate with bacteria found in tumor-susceptible mice and determine the aggressiveness of the microbiota.
Do these lipids directly impact intestinal bacteria?
We had to know whether the long chain fatty acids have a direct effect on the microbiota. We exposed cecal content of control mice to long chain fatty acids ex vivo in an approach applying biorthogonal non-canonical amino acid tagging (BONCAT) combined with fluorescence-activated cell sorting (FACS) that identifies translationally active bacteria. And bingo - this provided the mechanistic link showing that 6. ATF6-induced long chain fatty acids translationally activate specific bacteria including tumor-associated taxa, such as Desulfovibrio fairfieldensis. Not only was Desulfovibrio identified to be ATF6-associated in our TCGA data analyses, but we found Desulfovibrio fairfieldensis to be upregulated in tumor mice. Being a sulphate-reducing bacterium, we tested its capacity to produce the metabolite H2S (a known troublemaker in cancer) in response to long chain fatty acids. H2S production was increased, particularly in response the long chain fatty acid C22:0, providing mechanistic support for the tumor-promoting role of Desulfovibrio fairfieldensis.
The rest is history…
Following extensive revisions and rethinks (and probably a few existential crises) over a period of a year and a half, we wrapped up the story. We elucidated an evident triangular interplay between ATF6 signaling, lipid metabolism and the intestinal microbiota in the context of CRC. Why does this matter? We have moved a step forward in understanding what is going on in the intestinal epithelium following chronic ATF6 activation. This opens up an exciting avenue of research and presents a potential targetable vulnerability in CRC to extend prevention and treatment strategies. Targeting ATF6 or its lipid sidekicks could be a way to tweak the tumor microenvironment. Behind the scenes: Was it hard? Oh yes. Did experiments always work smoothly? Of course not – this is science. Was it fun and would we do it again? Absolutely.
Follow the Topic
-
Nature Metabolism

This journal publishes work from across all fields of metabolism research that significantly advances our understanding of metabolic and homeostatic processes in a cellular or broader physiological context, from fundamental cell biology to basic biomedical and translational research.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in