Unravelling the bioactive potential of a bacterial symbiont of a tropical bird
Published in Chemistry, Ecology & Evolution, and Microbiology
Why bacteria of the uropygial gland?
The uropygial (preen) gland of birds is located on the rump (on the dorsal side of the beginning of the tail), where it produces secretions that birds apply to their feathers. These gland secretions contain chemical compounds that allow for waterproofing and optimal feather cleaning. Multiple in silico studies have shown that these glands host diverse communities of bacterial symbionts, with potential antimicrobial activities, and thus might play crucial defensive roles. However, we realized that compared to other terrestrial systems, hardly any wet-lab experiments have yet been performed to validate sequencing data and identify the origin of bioactivities of these microbes.
The jungles of Papua New Guinea - the home of toxic birds
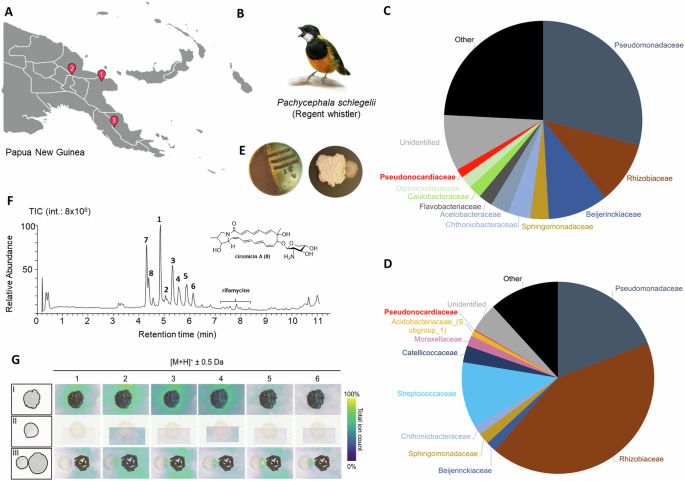
We (Knud Jønsson and Kasun Bodawatta from the University of Copenhagen) in collaboration with our colleges at the New Guinea Binatang Research Center (BRC) conducted our 2018 fieldwork in the YUS conservation area of Morobe province in Northern Papua New Guinea. The area was hardly reachable and only possible by using a 12-person charter flight that lands on a grass runway. In this pristine montane forest habitat, we studied different bird species presumed to be toxic, including Pachycephala schlegelii (Regent Whistler). Similar to poisonous frogs, birds have acquired the ability to consume toxic food and store the toxin in its feathers. While a few feathers were sufficient to analyze the content of the toxins, a study which was published early 2024, we also collected secretions of their uropygial glands for our microbiological studies. Only few microliters of the secretion were necessary to inoculate petri dishes with different media, such as potato dextrose agar medium with cycloheximide, to isolate bacteria. Together with our colleagues at BRC (Gibson Maiah) and the Papua New Guinea National Museum and Art Gallery (Bulisa Iova) we then - for the first time - analyzed the microbes that live within the uropygial glands of multiple individuals of P. schlegelii from different locations.
![Images of plane and the grassy runway at Yawan Village in YUS conservation area (top left), the field site (bottom right), field camp (bottom left) [Photo credits: Kasun Bodawatta], and an individual of Pachycephala schlegelii [photo credits: Ian Shriner] and a colony of Amycolatopsis PS_44_ISF1 [photo credit Elena Seibel]](https://images.zapnito.com/cdn-cgi/image/metadata=copyright,fit=scale-down,format=auto,quality=95/https://images.zapnito.com/uploads/UZygww6vSIquHbMhbCmQ_blog%20photo%201.png)
Is it a fungus or bacteria?
From our microbiological studies, we isolated multiple morphologically different colony forming units (CFU) of bacteria. One CFU was particularly interesting due to its antimicrobial activity. This filamentous colony was almost misidentified as a fungus, but visual observation and subsequent sequencing by our collaborator Michael Poulsen revealed that it was a bacterial strain belonging to the family Pseudonocardiaceae. To fully characterize the strain and its biosynthetic repertoire, it was necessary to generate a hybrid genome using Illumina and Nanopore sequencing technologies. A detailed genome analysis and comparison with genomes of other type strains uncovered that isolate PS_44_ISF1 belongs to an unclassified species of the genus Amycolatopsis. Interestingly, the isolate showed a relative close relation to another isolate Amycolatopsis sp. M39, which was isolated from an African fungus-farming termite, and additionally to A. saalfeldensis DSM44993, which was isolated from a former mine near Saalfeld in Thuringia (Germany).
Strong collaborations lead to novel discoveries
These findings were a peculiar coincidence as our collaborator Christine Beemelmanns, who at that time was leading a research group Chemical Biology of Microbe-Host Interactions at the Hans-Knöll-Institute in Jena (Thuringia, Germany), has intensively worked on both Amycolatopsis strains. Thus, Elena Seibel, a PhD student working on the metabolome of Amycolatopsis strains, then took a closer look at the bacterial strain and investigated its role within the birds microbiome and its natural product repertoire. First inhibitory assays of Amycolatopsis sp. PS_44_ISF1 showed moderate effects against fungal pathogens and feather degrading bacteria, suggesting a defensive symbiotic role of this bacterium. Intrigued by these findings, Elena wanted to find out which compounds may be responsible for the antimicrobial properties. With our high-quality genome sequence, Elena was able to first analyze in silco the encoded biosynthetic repertoire and which natural product family she might be able to detect in the metabolome. Elena quickly realized that the high quality genome data was essential for her analysis as the strain appeared to be a kind of “super producer” as to her surprise she found some similarities to her other Amycolatopsis strains, but also many unique genomic features. This allowed her to do a semi-targeted comparative metabolome analysis, which showed her that Amycolatopsis sp. PS_44_ISF1 indeed produces a series of antimicrobial compounds, including the macrolactam derivative ciromicin A and members of the rifamycin family. However, two yet unknown compound families caught our attention as they dominated the metabolome of strain PS_44_ISF1.
Hard work pays off
Following these preliminary findings, Elena, Soohyun Um and Tanya Decker started a high scale cultivation of the strain to isolate and characterize these metabolites. This included the cultivation and extraction of hundreds of plates, and a lot of patience during repetitive SPE and prep-HPLC runs to collect enough compound for NMR analysis. Finally, all the work was worth it as they discovered and structurally characterized seven new unreported metabolites. Six of these belonged to the same metabolite group (lipopeptides) and were named after the genus of the bird host (Pachycephala): pachycephalamides A-F (C33H62O10N8), while the 7th metabolite was named as demiguisin (C30H47N9O10) (the Harry Potter nerds among you will know where this name is originating from). The structural analyses of both compound classes turned out to become very challenging and required several isolation runs and different 2D NMR experiments and mass spectrometry-based fragmentation analysis. But Soohyun and Elena were up to the task and after months of isolation and analyzing NMR and MS-datsets, pachycephalamides turned out to be hybrid structures composed of a polyhydroxylated fatty acid or polyketide chain, and a peptide chain that could be derived from a non-ribosomal peptide synthase (NRPS). The structure of demiguisin turned out to be even more challenging, but at the end interpretation of the NMR data led us to conclude that the structure is a linear peptide composed of different and partially unnatural amino acids, which appeared to be heavily modified. Overall, the structural features of both compound classes appeared very uncommon and - to the best of our knowledge - were yet unreported structural features.
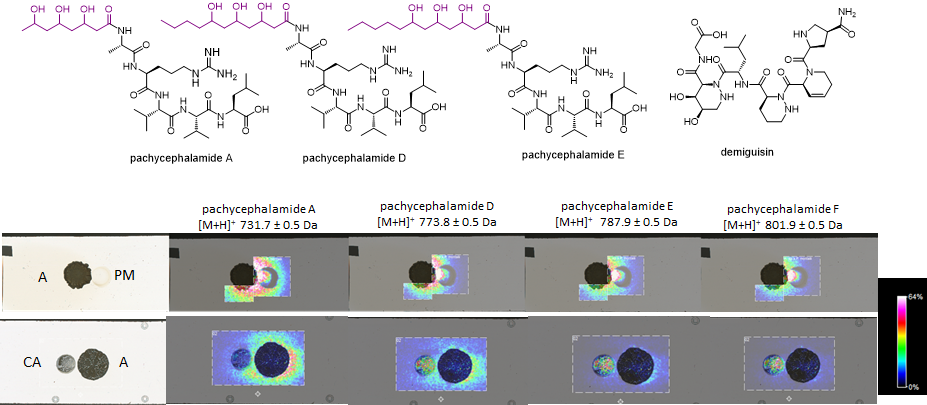
Bottom: MALDI Imaging showing a co-culture of Amycolatopsis sp. PS_44_ISF1 (A) and Pseudomonas monteilii (PM), and Candida albicans (CA). Intensity of molecular ion features of pachycephalamides are visualized and show an uneven distribution [picture credit @Anna Komor and Elena Seibel]
Following these metabolite discoveries and moving the laboratories to the Helmholtz Institute for Pharmaceutical Research Saarland, Elena also succeeded in analyzing the biosynthesis of these peculiar compounds. As the producer strain Amycolatopsis sp. PS_44_ISF1 turned out to harbour many different, and especially many fragmented gene clusters, Elena took the challenge to do an intensive analysis of bacterial transcriptomic data taken from conditions that either favored the production of the metabolites or inhibited their production. Finally, she was able to deduce about a dozen possible gene candidates, however, none of them perfectly fitted the collinearity assumption of a NRPS. It was not until Elena optimized knock out vectors and transformation protocols for this underexplored Amycolatopsis strain, that she was able to identify the biosynthetic gene clusters (BGCs) responsible for the production of these compounds. While these studies further supported the structural assignments of both compound classes, the characterization of the enzymatic machinery involved in the biosynthesis of these features will remain a big part of our future work.
Ecological and medicinal importance of novel peptides
Amycolatopsis sp. PS_44_ISF1 produces several antibiotics, including the macrolactam derivative ciromicin A and members of the rifamycin family. In addition, antimicrobial testing of purified compounds showed that the newly discovered pachycephalamides are tensioactive and thus have also antimicrobial effects against many known fungal and bacterial pathogens. Adding to the importance of pachycephalamides, we also found the ability of this compound to inhibit activity of different cysteine proteases, suggesting the potential for this compound to be useful as drug leads. Thus overall, we propose that the activity spectrum of the secreted metabolites aid in the overall sanitary effects of the birds feather. For this, we also validated the presence of Amycolatopsis in feather and uropygial gland microbiomes, and showed that pachycephalamides and demiguisin are present on some feathers of wild-caught P. schlegelii individuals. Overall the combined results support that these compounds may serve active antimicrobial roles in nature.
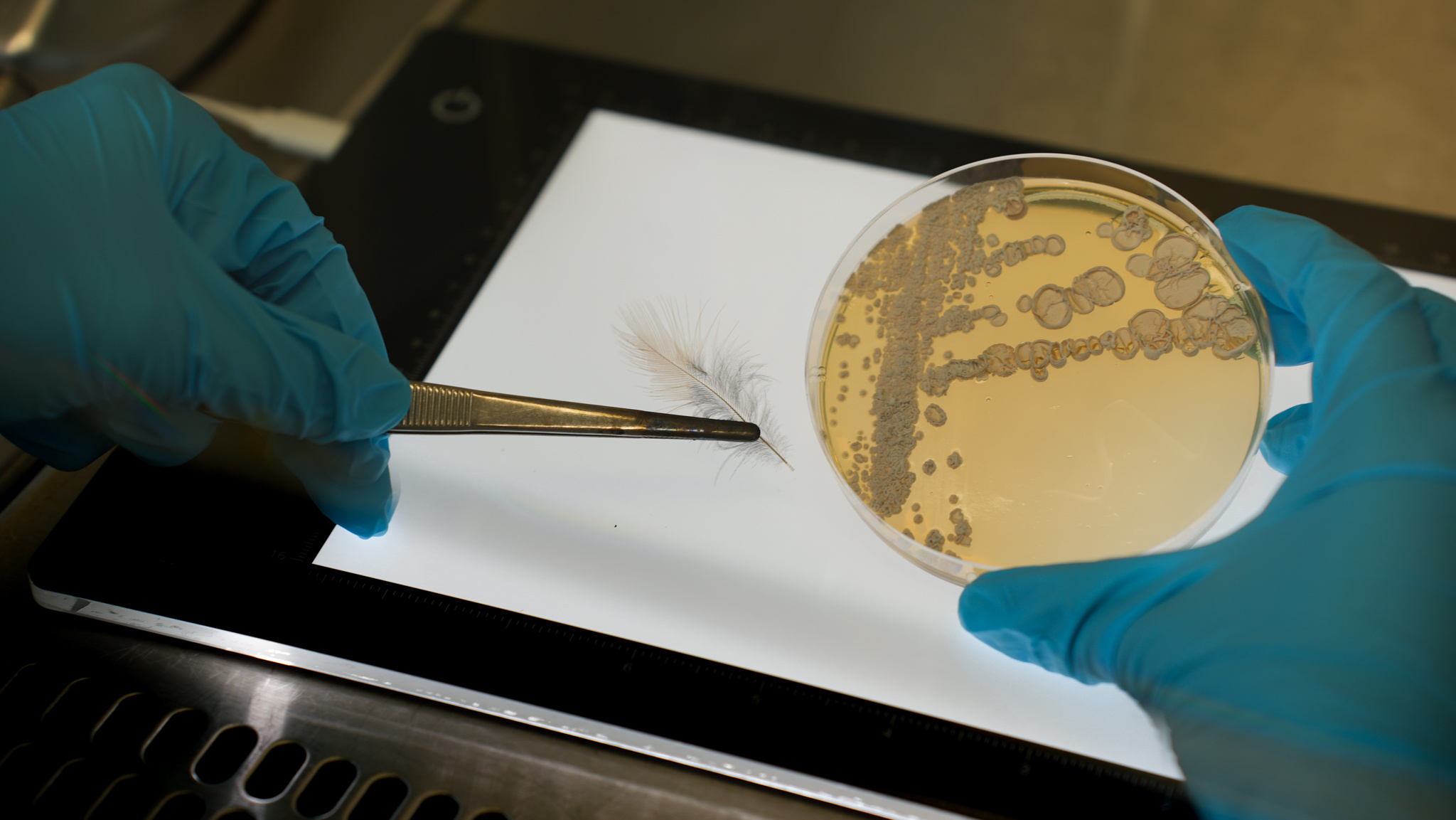
Representative picture of a birds feather (left) used for analyzing the presence and absence of protective metabolites and the birds toxin, and a plate culture (right) of the isolate Amycolatopsis sp. PS_44_ISF1 [photo credits:HIPS/Bernhardt].
The outcome of this study represents a great international collaborative effort, linking bird ecology with symbiotic interactions and natural product chemistry. Isolation of Amycolatopsis sp. PS_44_ISF1 from P. schlegelii and the discovery of novel bioactive compounds also supports that uropygial gland microbiomes harbor protective microbial species. Such discoveries are important as they can provide critical insights into the underlying mechanisms that facilitate symbioses between bird hosts and microbes. Although this study remains a small piece of a larger puzzle to disentangle the numerous unexplored symbionts of birds, it paves the way for discoveries of novel metabolites with both ecological and potentially medical relevance.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in