Unveiling the mystery of chromatin fibres
Published in Cell & Molecular Biology

Imagine the challenge of accommodating a 2-meter-long DNA string into a sphere with a mere 10-μm diameter while ensuring that each knot on the string is readily accessible in a time of need. This challenge is well met by chromatin, an intricate nucleoprotein complex that constantly transforms to support various fundamental life processes in the eukaryotic nucleus. Apart from storing the genomic information, chromatin plays pivotal roles in DNA replication, transcription, DNA damage repair, and more. Its structure lays down the very foundation for its function, determining the accessibility of the genome. Deciphering the elusive structure of chromatin is not only a pursuit driven by the natural curiosity of human but also crucial for understanding the above mentioned life processes which are closely associated with diseases, cancers, and aging.
Through decades of efforts, we now know that chromatin presents hierarchical structures with the building block nucleosome being compacted into the 10-nm beads-on-a-string structure that further coils into the so-called 30-nm chromatin fibre1. The fibre is believed to further condense to form larger chromatin domains in the nucleus. In chromatin, nucleosomes are connected by linker DNA which is regulated by the linker histone H12. The interconnection of nucleosomes and the trajectory of linker DNA are critical for determining the structure of chromatin fibres.
However, the existence of 30-nm chromatin fibres in the nucleus has been debated, as purified chromatin in moderate ionic conditions shows such structure under the electron microscope while it has not been supported in previous in vivo studies3-5. Moreover, various models for the 30-nm chromatin fibre have been proposed by different works but no consensus has been drawn. Two major models have been proposed: the zigzag and solenoid models6-9. In the zigzag model, nucleosomes zigzag back and forth with relatively straight linker DNA. While in the solenoid model, nucleosomes stack up linearly along the helical axis with heavily bent linker DNA. In both models, nucleosomes are compacted regularly with restricted distance and angle to the neighbouring nucleosome. Whereas recent studies employing single-nucleosome imaging suggests that nucleosomes are highly mobile and form heterogenous groups in chromatin10. The discrepancies among studies are largely due to the hypersensitivity of nucleosomes and chromatin fibres to the sample environment.
To ascertain the genuine structure of native chromatin fibres in the nucleus, we applied cutting-edge technologies cryo-focused ion beam (cryo-FIB) and cryo-electron tomography (cryo-ET) on the T-lymphoblast CEM cell. The cells were cryo-fixed in a hydrated state without any drastic perturbation, followed by stepwise thinning through cryo-FIB milling, yielding thin cell lamellae which are suitable for imaging by a transmission electron microscope. The very thin cell lamellae prepared in our work, 80-90 nm in thickness, gave rise to clear images of nucleosomes and chromatin fibres in intact nuclei (Fig. 1a). In some regions, we could even observe the linker DNA between nucleosomes with clear trajectories. Further analysis of the entire nucleosome population yielded a first high-resolution in situ structure of native nucleosomes and, more excitingly, identified two distinct classes of nucleosomes with or without linker histone H1 bound. Intriguingly, these two classes of nucleosomes exhibited a random distribution rather than a preferential one. These significant findings further enabled us to delineate the 3D organisation of nucleosomes within the chromatin fibres by placing individual nucleosomes back to the original tomograms (Fig. 1b). From the mapping back, we observed no rigid chromatin fibres. By measuring the distance and angle between adjacent nucleosomes, we found that the arrangement of nucleosomes within the native chromatin fibre was less constrained compared with the rigid models. Moreover, when dividing nucleosomes into subpopulations based on the distance and angle, we also identified short-ranged and partially compact di-, tri-, tetra-, and poly-nucleosomes in the chromatin fibre. Combining the direct visualisation, in situ structures, and population-wise statistical analyses, we reported a highly flexible and dynamic zigzag structure of chromatin fibres in the nucleus without uniformly compacted 30 nm fibres.
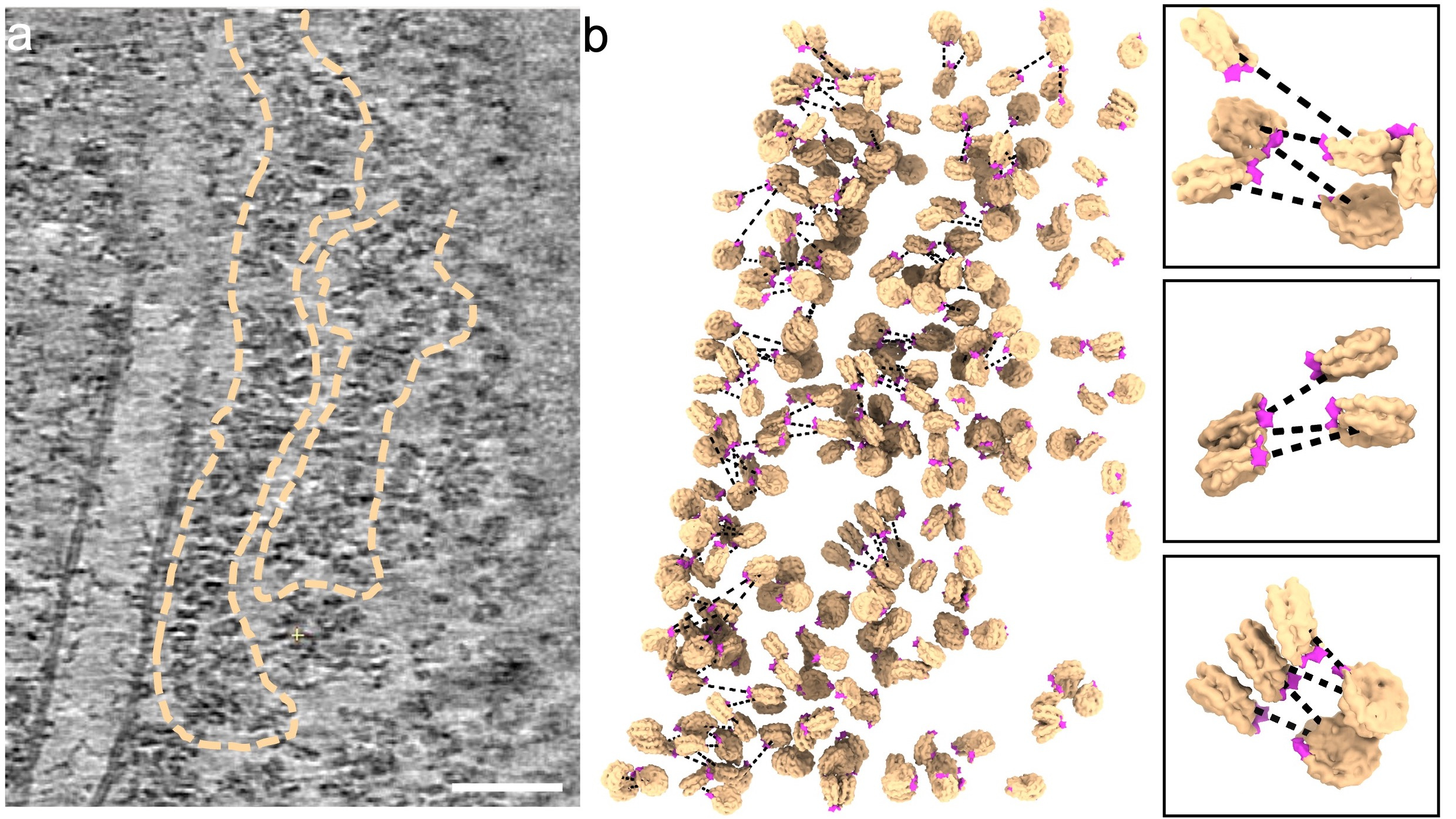
Figure 1 Organisation of nucleosomes in native chromatin fibres. a) A representative tomographic slice of chromatin fibres in the nucleus (from n = 5). Prominent fibre structures are indicated by dash outlines. Scale bar = 50 nm. b) Mapping of nucleosomes in chromatin fibres. Nucleosomes are mapped back to the tomogram based on their coordinates and orientations. The partial density of H1 is coloured magenta, indicating the entry and exit of the linker DNA, based one which, the DNA path (dashed lines) is predicted. Three sub-regions (circled) are showcased on the right panel.
This work, for the first time, provides unprecedented high-resolution insights into the structure of native chromatin fibres in intact nuclei, which not only answers the long-debated question of the chromatin fibre but also paves the way for future in situ investigations on the dynamic chromatin structure in response to cell division, gene expression and stresses.
References
- Olins, D.E. & Olins, A.L. Chromatin history: our view from the bridge. Nature reviews Molecular cell biology 4, 809-814 (2003).
- Thoma, F., Koller, T. & Klug, A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. The Journal of cell biology 83, 403-427 (1979).
- Eltsov, M., MacLellan, K.M., Maeshima, K., Frangakis, A.S. & Dubochet, J. Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proceedings of the National Academy of Sciences 105, 19732-19737 (2008).
- Cai, S. et al. Cryo-ET reveals the macromolecular reorganization of S. pombe mitotic chromosomes in vivo. Proceedings of the National Academy of Sciences 115, 10977-10982 (2018).
- Maeshima, K., Hihara, S. & Eltsov, M. Chromatin structure: does the 30-nm fibre exist in vivo? Current opinion in cell biology 22, 291-297 (2010).
- Schalch, T., Duda, S., Sargent, D.F. & Richmond, T.J. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature 436, 138-141 (2005).
- Finch, J.T. & Klug, A. Solenoidal model for superstructure in chromatin. Proceedings of the National Academy of Sciences 73, 1897-1901 (1976).
- Horowitz, R., Agard, D., Sedat, J. & Woodcock, C. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. The Journal of cell biology 125, 1-10 (1994).
- McGhee, J.D., Nickol, J.M., Felsenfeld, G. & Rau, D.C. Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell 33, 831-841 (1983).
- Iida, S. et al. Single-nucleosome imaging reveals steady-state motion of interphase chromatin in living human cells. Science Advances 8, eabn5626 (2022).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in