When cell biology meets ecology – uncovering the evolutionary and ecological context of a model bacterium
Published in Earth & Environment, Ecology & Evolution, and Microbiology
What is this research about?
Most research in bacteriology is strongly biased toward the study of a few selected model bacteria, leaving the vast majority of bacterial diversity unexplored. In this study, we addressed this knowledge gap by shedding light on the environmentally widespread but understudied bacterial order Caulobacterales. Although this order includes the well-characterized model organism Caulobacter crescentus, our knowledge of its diversity and ecology has remained limited. By integrating phylogenetic, comparative genomic, cell biological, and ecological approaches, we reveal a previously unrecognized diversity within this group, which provide new insights into the evolution of fundamental bacterial features.
How did it all start?
For over 13 years, my lab has focused on investigating the molecular mechanisms that regulate bacterial growth and cell cycle progression in response to changing environmental conditions. Our primary model organism during this time has been Caulobacter crescentus, an asymmetric bacterium with a dimorphic lifecycle that has emerged as one of the prime models for bacterial cell biology. While extensive research has clarified many of the molecular mechanisms behind its unique lifecycle, surprisingly little is known about its ecology and evolutionary context. For example, it remains unclear in which environments it occurs, who its closest relatives are, and how representative the characteristic cell biological features of C. crescentus are for the entire order.
Just as I recruited a new PhD student, Joel Hallgren, to explore some of these questions, I met Sarahi Garcia. We both were Fellows at the Science for Life Laboratory, a national center for life sciences research in Sweden. Sarahi, whose background is in microbial ecology, had recently started her own lab to investigate microbial interactions in freshwater habitats. In her group, they had compiled a large collection of public metagenomic datasets and were in the process of analyzing them. We connected at a time when both of us were open to new research directions. We had flexible funding not tied to specific projects and had recently recruited new members to our labs. When Sarahi mentioned to Joel and me that her datasets and her newest cultivated communities included numerous Caulobacter genomes, it caught our attention. We began to wonder whether her datasets and ecological expertise could help us address some of the open questions regarding Caulobacter biology.
What was the research process like?
We held a series of initial meetings to discuss our research and identify potential areas of common interest. We were full of ideas but also realized that we were speaking different scientific “languages”, coming from fields that use very different approaches and ways of addressing research questions. At first, it was still uncertain which questions we should focus on, how to approach them, and what specific roles each of us would play in the collaboration. My lab had extensive knowledge of the Caulobacter model organism and long-standing experience with experimental work, but we lacked expertise in handling large datasets and performing phylogenetic analyses. In contrast, Sarahi’s lab had strong experience with bacterial genome data. Through her postdocs Julia Nuy and later Jennah Dharamshi, her group also acquired deep expertise in phylogenetics, which was critical for our plans to map out the diversity of Caulobacterales members.
Ultimately, it was Joel’s curiosity and open-mindedness—combined with Jennah’s (and initially Julia’s) high technical expertise and motivation to contribute—that got the project off the ground. Their joint effort led to the first discoveries and allowed a coherent story to begin to take shape. We did not initially plan for Joel to work on the bioinformatics aspects of the project, but he found it so rewarding to combine bioinformatics-driven questioning with experimental cell biology that he took programming courses and ended up making it his main PhD thesis project.
What did we find?
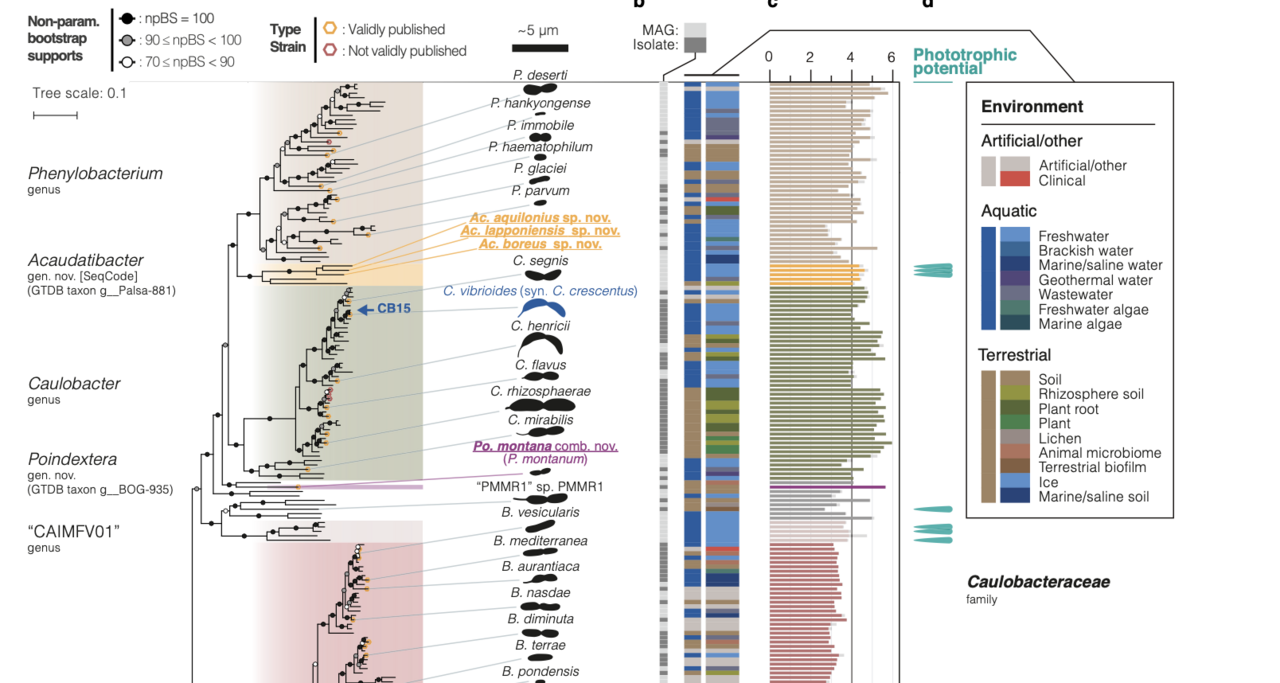
We began by constructing an updated phylogeny that included around 350 representative species, including a significant number of metagenome-assembled genomes from uncultured bacteria. By extracting sampling-site information from the associated metadata, we were able to map the environmental distribution of these species. This updated phylogeny enabled several important taxonomic corrections and, more importantly, provided a solid foundation for investigating the diversity, evolution, and ecology of Caulobacterales members.
Among our most striking and unexpected findings was that two independent lineages had lost a large set of approximately 100 genes associated with cellular dimorphism—an exceptional example of how lifecycle complexity can be reduced to a simpler form. We also discovered that a significant subset of species possessed genes related to phototrophy, including one lineage, belonging to a new genus that we named Acaudatibacter, which possesses the complete set of genes for full photosynthesis—an entirely unrecognized feature of the Caulobacterales. Finally, our analyses shed new light on the evolutionary history of crescentin, suggesting that this signature Caulobacter cytoskeletal protein may have originated from a bacteriophage.
How will this research continue?
The research opens numerous new directions for future investigation. As one of the reviewers noted, it “forms the basis for years of experimental work ahead.” For example, our study identified a set of candidate genes that may represent new players involved in dimorphism and thus deserve further characterization. Our findings provide a foundation for better understanding the evolution of lifecycle complexity, and the discovery that closely related photosynthesis genes are widespread among the Caulobacterales suggests that their common ancestor may have been phototrophic. Finally, given that some of the most intriguing features were found in bacteria that remain uncultured, our work underscores the need to isolate and characterize additional strains to uncover novel biological insights beyond the well-studied model organisms.
Lessons learned
We believe that our study highlights the power of interdisciplinary research. Although such collaborations can be challenging at times, they often open new research directions that would not have emerged within a single research group alone. Generous long-term funding was essential for pursuing this path, together with a team of open-minded lab members who are passionate about science and courageous enough to step outside their comfort zones.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Your space to connect: The Myeloid cell function and dysfunction Hub
A new Communities’ space to connect, collaborate, and explore research on Clinical Medicine and Cell Biology!
Continue reading announcementRelated Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in