How pancreatic cancer taught us where it plans to go
Published in Cancer
When our team first gathered around a whiteboard in the lab at Columbia, we were staring at a simple question that had bothered clinicians for years: Why do some pancreatic cancers head straight for the liver while others prefer the lung? The survival differences are enormous, but the field had no satisfying explanation. It was one of those mysteries everyone talks about in the hallways but no one has the data to crack open.
Pancreatic ductal adenocarcinoma, or PDAC, is already a brutal disease. Most patients who are lucky enough to have surgery eventually see their cancer come back. But as we combed through records of 744 patients who had undergone resection at our center, a stark pattern emerged: patients whose cancer returned first in the liver lived dramatically shorter lives than those whose disease recurred in the lung. We were looking at the same diagnosis, the same organs, the same surgeries, the same treatments. Yet the trajectories split sharply, almost as if the tumors were following two very different scripts.
That observation lit the fuse. If these patients entered the operating room with no visible metastases, maybe the primary tumor itself already held clues. Maybe, inside the mass removed at surgery, there were hidden “states” of cancer cells quietly preparing for the journey ahead, already wired to thrive in one organ or another.
But PDAC is notoriously hard to profile at single-cell resolution. The tissue is dense and fibrotic, almost stone-like, and traditional single-cell RNA-seq methods tend to shred it. We needed a workaround. After months of troubleshooting, test runs, and more than a few late-night Slack messages, our team turned to a newer approach: single-nucleus RNA sequencing combined with low-pass whole genome sequencing, applied directly to small archival samples. Suddenly, the door cracked open.
From 21 primary tumors we pulled out nearly 92,000 high-quality nuclei. When we separated malignant from nonmalignant cells and looked at their transcriptional states, something unexpected emerged: cancer cells from tumors that later metastasized to the liver already expressed genes that liver cells normally use. And cancers that would eventually travel to the lung? Their primary tumors were quietly echoing lung epithelial programs.
It was as if the cells were rehearsing for their future home.
At first, we didn’t quite believe it. Maybe this was an artifact of tissue processing. Maybe it reflected general “hydration signatures,” or tumor subtype differences, or DNA-level alterations. So we threw every validation test we had at it. We checked metastases directly. We evaluated spatial transcriptomes. We scanned scRNA-seq data from primary liver and lung cancers. We even tapped into MetMap500, a massive atlas of how human cancer cell lines metastasize in mice.
Each dataset told the same story: these organ-specific transcriptional signatures weren’t accidents. They marked real biological states that predicted where PDAC cells were headed.
And then came the twist. When we used Echidna, a computational tool that measures how much gene expression is driven by underlying copy number changes, we found that the organotropic signatures were not explained by genomic alterations. The DNA wasn’t telling the cells to behave this way. It was something else: gene transcriptional reprogramming. The cancer cells were borrowing the identity of the organ they were preparing to invade; those transcriptionally resembling liver cells (hepatocytes) eventually metastasized to the liver, while those resembling lung cells (bronchoalveolar) metastasized to the lung (see the video)!
That realization reframed everything. PDAC wasn’t just evolving randomly; it was adopting organ-like programs early, maybe even at premalignant stages. When we reconstructed tumor development from acinar-to-ductal metaplasia through PanIN to full-blown cancer, those future metastatic programs were already faintly visible. The tumor microenvironment shaped some of this, too. Liver-tropic tumors showed signals of IGF1-driven metabolic conditioning, while lung-tropic ones carried interactions supporting a bronchoalveolar-like state and distinct immune ecosystems.
By the time we stepped back, the picture had become surprisingly clear: organ-specific recurrence in PDAC is not an accident of late-stage spread. It’s written into the biology of the primary tumor long before metastasis is clinically detectable.
And for patients, that matters. The first recurrence site isn’t simply a waypoint; it predicts survival, treatment needs, and clinical outcomes. If we can detect these transcriptional programs early, we may eventually tailor surveillance, personalize therapy, and identify intervention points before metastatic destiny becomes metastatic reality.
For us as a team, this project felt like assembling a puzzle with pieces from genomics, pathology, computational biology, clinical oncology, and good-old-fashioned persistence. Along the way, we were humbled by how deeply a cancer can mimic the tissues around it, even those it hasn’t reached yet. And we came away with a sense that organotropism, once treated as a black box, is finally becoming something we can understand, measure, and maybe one day disrupt.
This paper was many years in the making. But the idea that cancers “speak the language” of the organs they plan to colonize has stayed with all of us. And we hope this work nudges the field closer to predicting metastasis not as a surprise, but as a decipherable biological trajectory.
Follow the Topic
-
Nature Genetics

This journal publishes the very highest quality research in genetics, encompassing genetic and functional genomic studies on human and plant traits and on other model organisms.
Ask the Editor – Inflammation, Metastasis, Cancer Microenvironment and Tumour Immunology
Got a question for the editor about inflammation, metastasis, or tumour immunology? Ask it here!
Continue reading announcement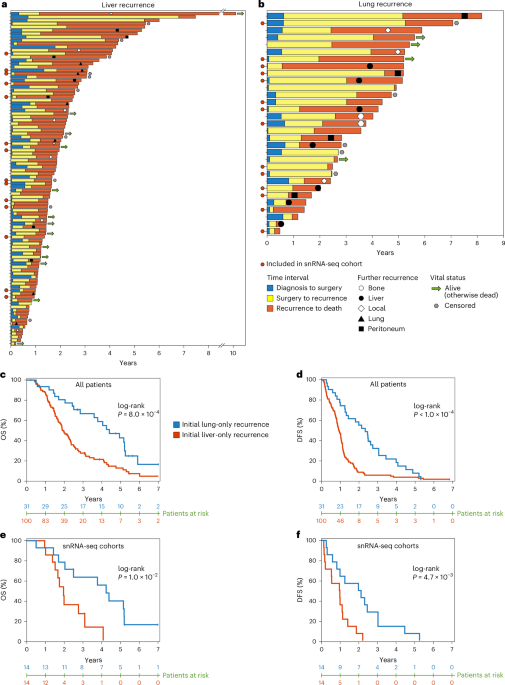

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in