A foundation model for enhancing magnetic resonance images and downstream segmentation, registration and diagnostic tasks
Published in Protocols & Methods, Computational Sciences, and Anatomy & Physiology
Challenges in Structural MRI
Structural magnetic resonance imaging (MRI) is a well-established, non-invasive, and safe technique for characterizing the human brain, valued for its excellent soft tissue contrast and absence of radiation. However, despite these advantages, participant movement—whether due to head motion, heartbeats, breathing, or even blinking—during image acquisition can introduce significant blurring and ghosting artifacts. These artifacts create confounding effects, complicating subsequent neuroimaging analyses, such as tissue segmentation, registration, atlas construction, parcellation, and group comparisons.
Motion artifacts are prevalent across all age groups, from fetuses to elderly adults. Collecting high-quality MRI from young children (2–4 years of age) is particularly challenging, as they are highly active and often struggle to remain still during scans. This leads to a success rate of only 33%–60% for usable scans, as reported in the literature. Beyond motion artifacts, brain MRI is frequently acquired with large slice thicknesses or low resolution due to hardware limitations, signal-to-noise ratio constraints, scan time restrictions, and the need to ensure patient comfort. An additional challenge in utilizing MRI data is the significant inter-center variability, driven by differences in scanner manufacturers, models, and imaging protocols. This heterogeneity limits cross-center comparability and negatively impacts both clinical and research applications.
BME-X Model
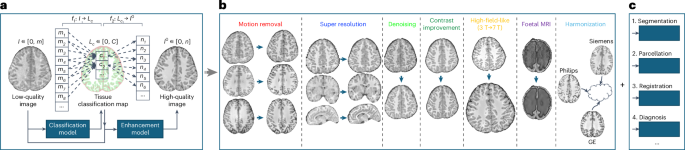
To address these challenges and enhance brain MRI quality through motion correction, super-resolution, denoising, harmonization, and contrast enhancement, we propose a flexible and easy-to-implement brain MRI enhancement foundation model, BME-X, as shown in Fig. 1. The core assumption behind BME-X is that improving image quality results in clearer, sharper images with consistent intensity ranges across tissue types. This simplifies the complex task of image reconstruction into a tissue classification problem, enabling the model to estimate high-quality images guided by classification results.

Figure 1. Overview of the brain MRI enhancement foundation (BME-X) model. a, Flowchart of the tissue-aware foundation model to improve image quality. b, Seven applications of the foundation model, including motion removal, super-resolution reconstruction, denoising, contrast improvement, high-field-like reconstruction, enhancement for fetal MRIs, and harmonization. c, Extensions for downstream tasks, including tissues segmentation, parcellation, registration, and diagnosis.
The BME-X model includes two key components: a tissue classification network and a tissue-aware enhancement network. The tissue classification network is trained to predict tissue labels, leveraging deep learning-based methods for robust classification of motion-corrupted MR images. The tissue-aware enhancement network then uses these labels to generate high-quality, enhanced images. Moreover, our harmonized, high-quality, high-resolution images generated by BME-X facilitate various downstream tasks including tissue segmentation, parcellation, registration, and diagnosis, as evidenced by our experimental results.
Applications
Extensive testing on over 13,000 images from diverse datasets demonstrates that BME-X outperforms state-of-the-art methods in motion correction, super-resolution reconstruction, denoising, handling pathological MRIs, and harmonizing images across Siemens, GE, and Philips scanners. Figure 2a illustrates the age distribution of the 10,963 testing images, which were acquired using various scanners (Siemens, GE, and Philips) and cover a wide age range, from fetuses to the elderly. Figure 2b showcases representative lifespan images, while Figure 2c highlights tissue contrast t-score (TCT) values before and after enhancement for the 10,963 in vivo testing images. The results show a significant improvement in image quality after enhancement (two-sided t-test, P < 0.001), as indicated by higher TCT values. Figure 3 demonstrates BME-X’s impact on reducing scanner variability. The top section (blue) presents in vivo images from Siemens, GE, and Philips scanners at 1.5T and 3T, emphasizing differences in imaging parameters and dynamic intensity histograms. The bottom section (orange) displays the enhanced images, which exhibit more consistent contrast and uniform histograms, underscoring the model’s effectiveness in harmonizing disparities caused by diverse acquisition protocols.

Figure 2. Enhanced results of the BME-X model for 10,963 in vivo low-quality images across the whole human lifespan, collected from 19 datasets. a, Age distribution. b, Original images and the corresponding enhanced results. c, Comparison of TCT values for the 10,963 original images and the corresponding enhanced images by BME-X.
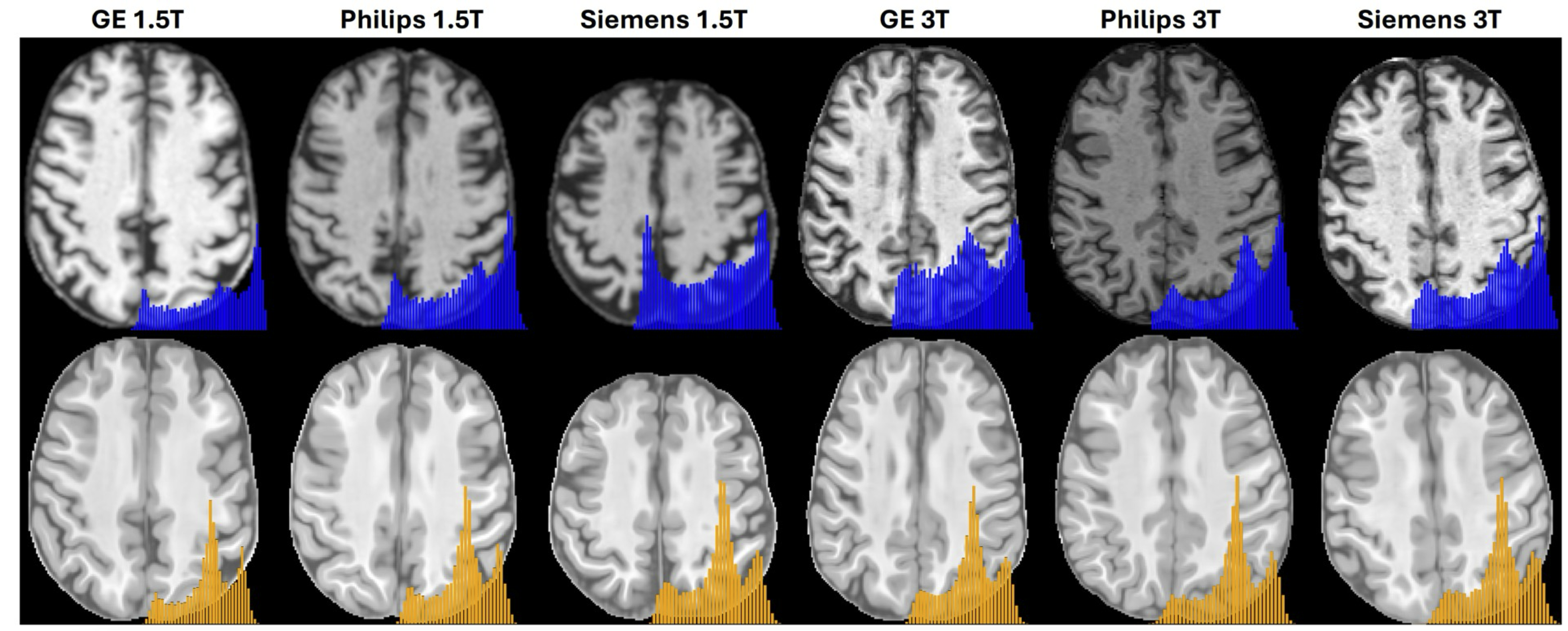
Figure 3. Application on harmonization across scanners (into a latent common space). The top section (blue) shows the original images acquired using GE, Philips, and Siemens scanners with 1.5T and 3T field strengths, each employing different imaging parameters. These variations introduce significant disparities in image contrast and appearance, resulting in dynamic histograms. The bottom section (orange) presents the corresponding enhanced images. Following enhancement with the BME-X model, the histogram distributions demonstrate significantly improved consistency.
Advancing Neuroimaging
The implications of our findings are profound for both human health and neuroimaging research. By significantly enhancing brain MRI quality across diverse populations and scanner types, the BME-X model enables more accurate and reliable analysis of brain structures, which is crucial for the early detection, diagnosis, and monitoring of neurological conditions. Enhanced image quality, particularly in challenging cases involving motion artifacts or low-resolution scans, facilitates clearer visualization of brain anatomy and pathology, potentially leading to better-informed clinical decisions.
Follow the Topic
-
Nature Biomedical Engineering

This journal aspires to become the most prominent publishing venue in biomedical engineering by bringing together the most important advances in the discipline, enhancing their visibility, and providing overviews of the state of the art in each field.
Related Collections
With Collections, you can get published faster and increase your visibility.
Implantable wireless communication technologies
Publishing Model: Hybrid
Deadline: Nov 28, 2026
Biosensing
Publishing Model: Hybrid
Deadline: Jun 30, 2026

Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in