A marine viral halogenase that iodinates diverse substrates
Published in Chemistry

Our search for wildly different wild type halogenases:
https://www.nature.com/articles/s41557-019-0349-z
In 2004, when my research team started working with halogenases, there were very few flavin-dependent halogenases (FDHs) known and characterised. Even today, this has remained the case, though a series of FDHs are well characterised, this portfolio has remained very focussed on tryptophan (PrnA, RebH, ThaI, KtzQ, PyrH, KrmI etc) with a few other exciting exceptions.1-3There has been exquisite work on engineering and evolving several of these FDHs,4, 5but with >5000 structurally diverse natural halometabolites6we felt that some wildly different, naturally evolved halogenases must be out there. One has just to be able to predict their function on a sequence level.

A viral halogenase that can iodinate a broad palette of substrates in the laboratory
Danai Gkotsi, then a PhD student in the group, in a collaborative project with Syngenta, carried out a careful comparative analysis of every halogenase that had been characterised in vitro. Firstly, we looked for conserved sequence motifs and found a new fingerprint that appeared in every single halogenase. We could use this fingerprint by itself to identify halogenases in sequence databases. We wanted to look at organisms that were phylogenetically very different to those where the original halogenases were found, so when we saw the sequence within a cyanophage whose hosts are the most abundant photosynthesisers on the planet, we decided to take a closer look.
We heterologously produced the enzyme-VirX1 (named so because it is the first biochemically and structurally characterised halogenase from a virus), and screened its activity against a substrate library. From our initial experiments, we thought that perhaps our halogenase had no activity – but when we switched from sodium chloride, to bromide and iodide salts we were stunned to see that it had a preference for iodination and is capable of halogenating a very broad range of substrates.
Danai and Hannes Ludewig (a joint PhD student between the Naismith and Goss groups), were able to crystallise the enzyme, revealing its strikingly open structure. A structurally diverse range of compounds were seen to be good to excellent substrates, and certain iodo-products were characterised by Sunil Sharma, a postdoc within our group. A series of challenging spirocyclic compounds were obtained from our collaborators in York (Will Unsworth and Richard Taylor) and we were thrilled to see that our enzyme could regioselectively halogenate several of these.
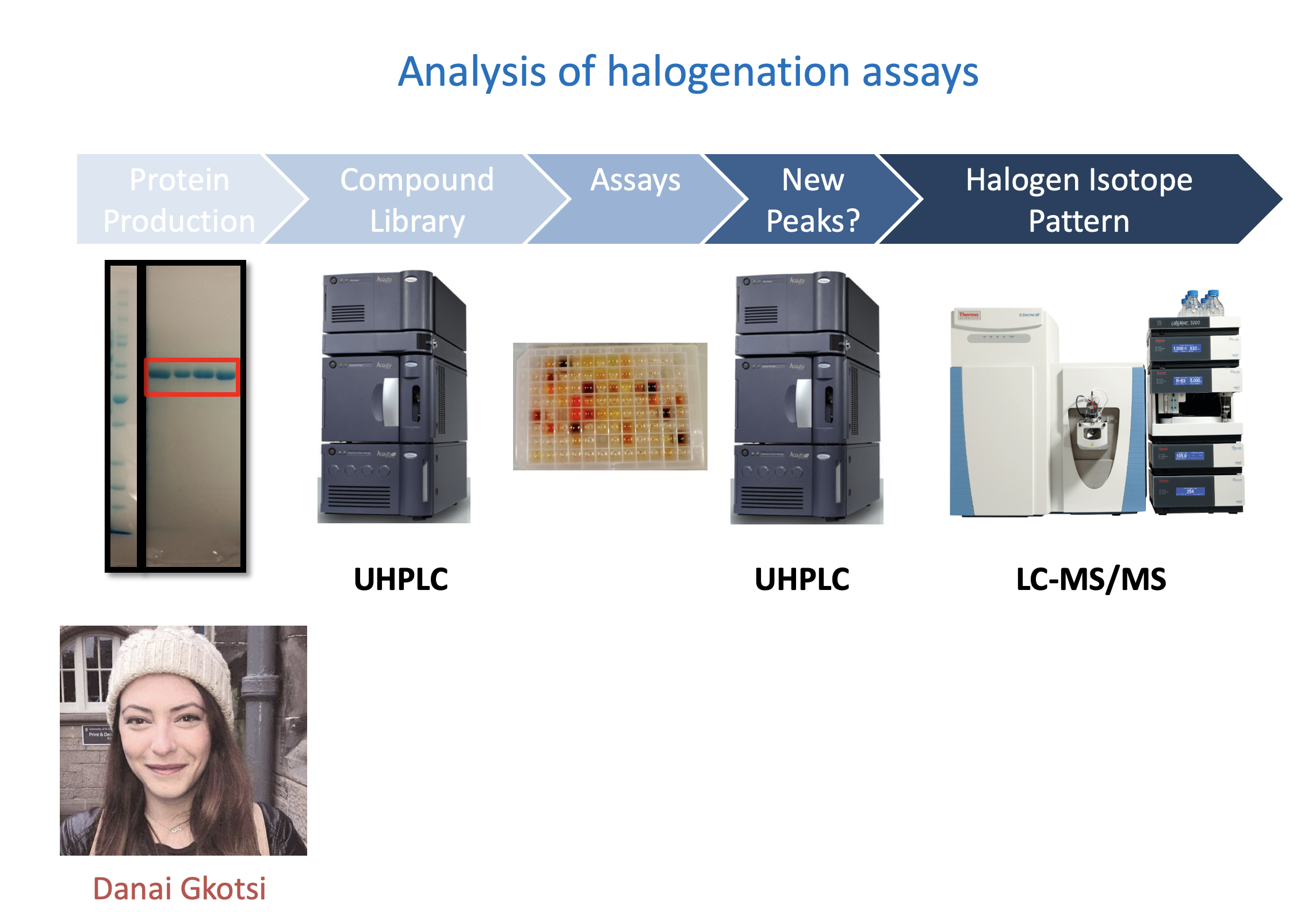
Workflow for halogenase testing
We had been looking for tools for our GenoChemetics approach to Natural Product generation https://doi.org/10.1021/ja1060406 and are really excited to now have this gene in hand that enables late stage halogenation/activation of complex structures. We are also working in partnership with industry to determine how this and other enzymes may be harnessed as tools for selective and green halogenation of pharmaceuticals, and high-value intermediates. And what is the natural role of this halogenase within the virus? This is a question that we are working with collaborators to address.
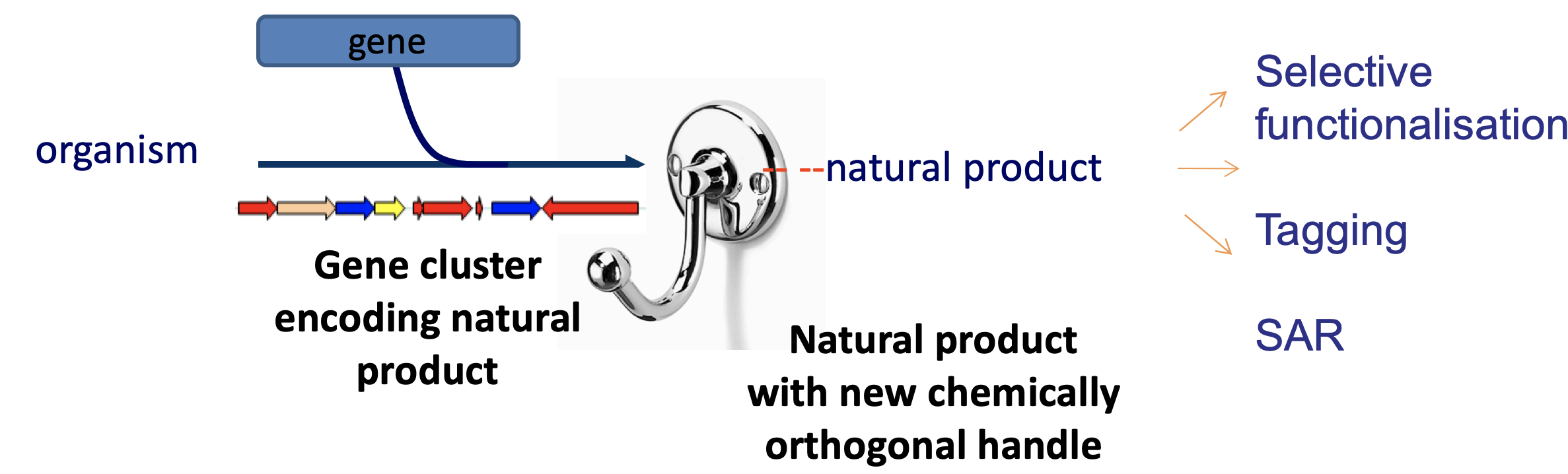
A new tool for selective and late stage C-H activation as well as a potential new tool for the GenoChemetic modification of natural products https://doi.org/10.1021/ja1060406
https://www.nature.com/articles/s41467-017-00194-3
REFERENCES
For example see:
6) https://doi.org/10.1002/anie.201408561
7) https://doi.org/10.1021/ar9701777
Follow the Topic
-
Nature Chemistry

A monthly journal dedicated to publishing high-quality papers that describe the most significant and cutting-edge research in all areas of chemistry, reflecting the traditional core subjects of analytical, inorganic, organic and physical chemistry.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in