A step towards synthetic life
Published in Bioengineering & Biotechnology

Synthetic biology encompasses the efforts of a broad and interdisciplinary scientific community to manipulate living matter at an unprecedented level of control and complexity. In this context, bottom-up synthetic cells and organelles are designed with defined functionalities in mind and assembled from natural or man-made building blocks. To a large extent, the current efforts are focused on understanding the principles of life, that govern the interactions between these building blocks. Therefore, present designs are mostly aiming to reproduce or mimic essential cellular processes, such as growth, motility, metabolism and reproduction. Thereby, these bottom-up replicated processes are built and developed as separate modules, and then integrated in synthetic constructs in a step-wise manner. Notably, regardless whether being natural or synthetic, these processes are usually out-of-equilibrium, hence they require continuous supply of energy, quite often in the form of adenosine triphosphate (ATP). Consequently, energy regeneration modules are essential components of virtually every synthetic construct.
In the Max Planck Institute for Dynamics of Complex Technical Systems we design and construct such energy regeneration modules based on photophosphorylation and oxidative phosphorylation, and also develop approaches to integrate those with other processes1,2. This work started under the umbrella of a large network initiative on Bottom-up Synthetic Biology within the Max Planck Society (MaxSynBio), which gave a significant research momentum and is continuously seeding other projects and organizational settings, such as the recently founded Max Planck School Matter to Life. In our modules, ATP is synthesized by a complex transmembrane enzyme (ATP synthase) inserted in the membranes of nano-sized vesicles. The synthesis is driven by a proton gradient (difference in concentrations across the membrane), which is established and maintained via the proton translocation activity of co-inserted proton pumps. A significant disadvantage of such synthetic phospholipid vesicles equipped with various membrane proteins is their low functional and structural durability. In the absence of the repair and replacement mechanisms existing in nature, the inserted enzymes are unstable and lose their activity. To address this, we previously successfully inserted the energy regeneration protein machinery into polymer compartments made of a common emulsifier (PDMS-g-PEO)3, which is miscible with lipids as well as with block copolymers, thus enabling the formation of the so-called hybrid vesicles. Notably, PDMS-g-PEO forms membranes with a thickness and fluidity similar to natural membranes, thus presenting an enzyme-compatible environment. In our recent study4, we were able to extend the functional lifetime of a bacterial proton pump to nearly two weeks by enzyme integration into PDMS-g-PEO-based membranes. Furthermore, the polymer also protected the enzymes against oxidative damage.
With the energy modules stabilized and their functionality increased, we are now exploring avenues for their expansion and refinement, for instance via insertion of additional enzymes and better control over their orientation. To this end, we were looking to replicate a highly dynamic process of membrane remodeling through fusion in the otherwise static polymer membranes. We intentionally did not employ one-off physiochemical triggers such as electrostatic attraction, DNA-mediated fusion and others because we were aiming to construct a fusion platform, which would enable directed and sustained fusion, while being as biocompatible and versatile as possible. This required us to expand the building block toolbox with remarkable new tools – SNARE fusogenic peptides. We teamed up with the group of Reinhard Jahn, who is a renowned expert on the latter and reconstituted SNAREs in polymer and hybrid vesicles with outward-facing (fusion-promoting) orientation (Fig. 1).
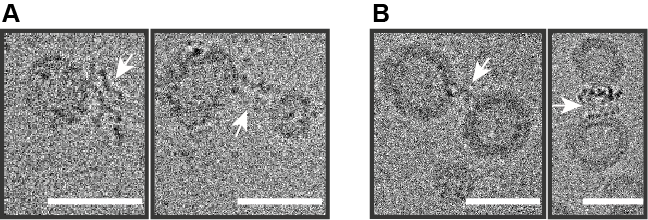
Fig. 1. Reconstitution of SNAREs into polymersomes and hybrids. SNAREs, indicated with white arrows, were inserted into polymer (a) and hybrid (b) vesicles via detergent removal by size exclusion chromatography. Scale bars = 30 nm.
Next, we examined the SNARE-mediated membrane and content mixing. Remarkably, SNAREs were able to provide sufficient energy to trigger prompt and efficient full fusion of man-made membranes, which we demonstrated through integration of two respiratory enzymes towards a coupled energy regeneration module (a minimal respiratory chain), whereby the activity of the module in polymer membrane surpassed the one determined in their natural counterparts (Fig. 2).
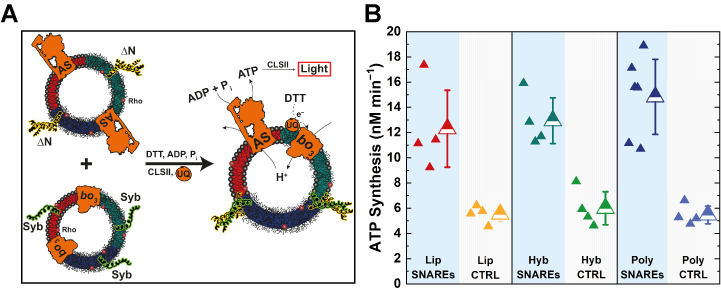
We then joined forces with the teams of Reinhard Lipowsky, Rumiana Dimova and Panagiotis Kastritis to determine and describe the set of conditions that governed the successful SNARE-mediated fusion of polymer membranes. We are excited to see how the charted parameters can be applied to different types of artificial membranes with the aims of identifying unexpected synergies between natural and man-made material. Meanwhile, we are hoping that the outlined fusion intermediates will serve as a solid starting point on the road to understanding SNARE-induced remodeling of polymersomes, thus paving the way for future endo/exocytosis biomimetics.
Related publication:
https://www.nature.com/articles/s41467-021-25294-z
Associated publications:
1. Otrin, L. et al. Artificial Organelles for Energy Regeneration. Advanced Biosystems 3, 1800323, doi:10.1002/adbi.201800323 (2019).
2. Ivanov, I. et al. Bottom-Up Synthesis of Artificial Cells: Recent Highlights and Future Challenges. Annual Review of Chemical and Biomolecular Engineering 12, 287-308, doi:10.1146/annurev-chembioeng-092220-085918 (2021).
3. Otrin, L. et al. Toward Artificial Mitochondrion: Mimicking Oxidative Phosphorylation in Polymer and Hybrid Membranes. Nano Letters 17, 6816-6821, doi:10.1021/acs.nanolett.7b03093 (2017).
4. Marušič, N. et al. Constructing artificial respiratory chain in polymer compartments: Insights into the interplay between bo3 oxidase and the membrane. Proceedings of the National Academy of Sciences 117, 15006-15017, doi:10.1073/pnas.1919306117 (2020).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in