All TP53 mutations are NOT equal: Does the "deleterious effect" of the TP53 mutation affect the outcome of Myelodysplastic Syndrome?
Published in Cancer

Full paper on Blood Cancer Journal website
What was the question?
 All TP53 mutations are NOT equal. TP53 mutations are diverse and distributed across the codons of the entire coding region. Different types of TP53 mutations lead to distinct functional consequences such as oncogenic gain-of-function and protein loss-of-function with dominant-negative effect. Recent studies in myelodysplastic syndromes (MDS) have identified specific mutational and cytogenetic characteristics that are associated with improved outcomes: low variant allele frequency (VAF), mono-allelic TP53 alterations, and absence of complex or monosomal karyotype (CK/MK). However, the impact of the different types of TP53 mutations per se on the phenotype and outcome in MDS is still largely unknown.
All TP53 mutations are NOT equal. TP53 mutations are diverse and distributed across the codons of the entire coding region. Different types of TP53 mutations lead to distinct functional consequences such as oncogenic gain-of-function and protein loss-of-function with dominant-negative effect. Recent studies in myelodysplastic syndromes (MDS) have identified specific mutational and cytogenetic characteristics that are associated with improved outcomes: low variant allele frequency (VAF), mono-allelic TP53 alterations, and absence of complex or monosomal karyotype (CK/MK). However, the impact of the different types of TP53 mutations per se on the phenotype and outcome in MDS is still largely unknown.
To assess this, we quantified the deleterious effects of missense TP53 mutations using a computationally-derived evolutionary action score (EAp53, range, 0-100, a higher score indicates worse impact) in 270 newly-diagnosed TP53-mutant MDS patients treated with hypomethylating agents and correlated with clinicopathologic and genomic features.
What were the findings?
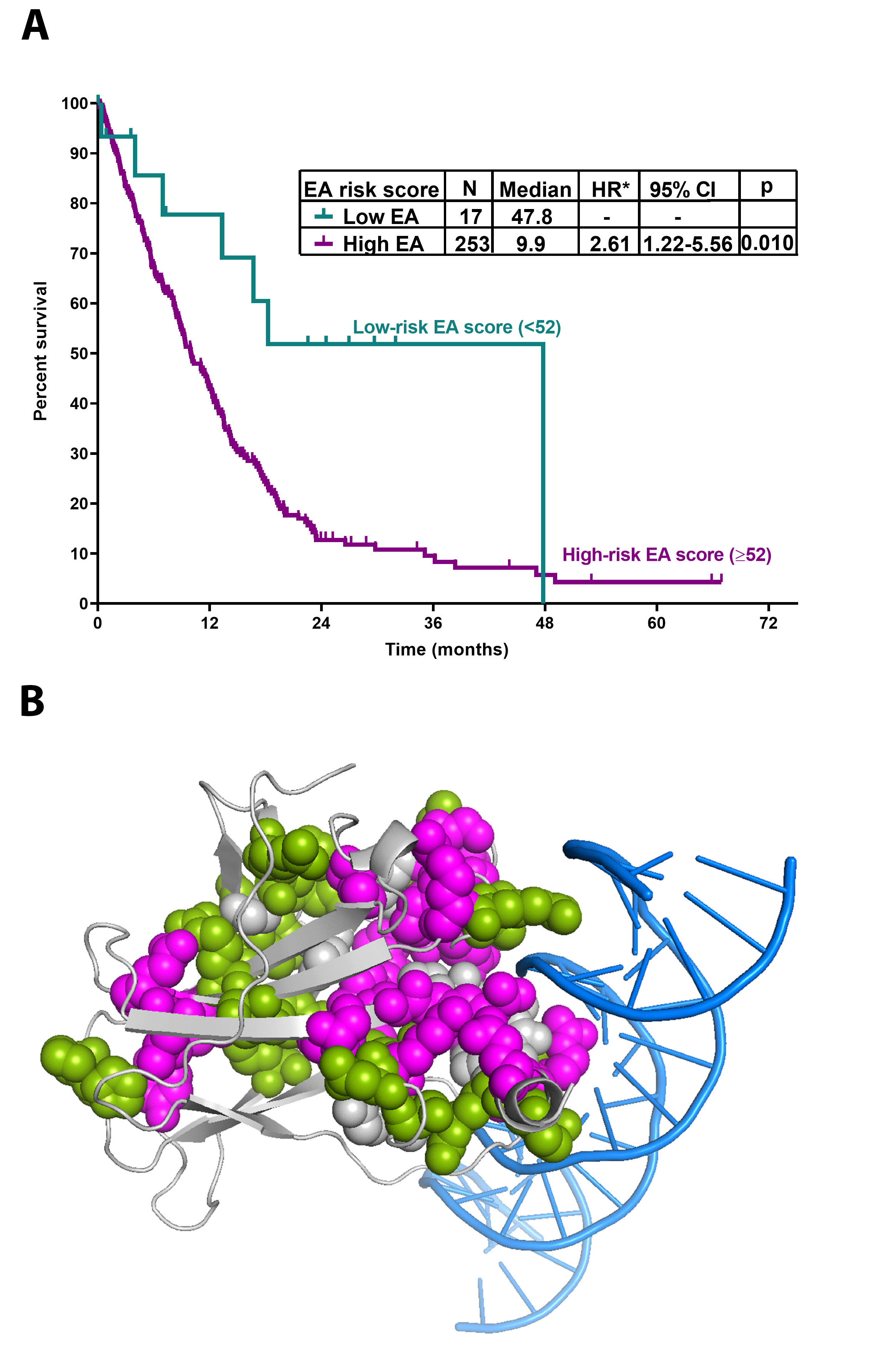
We found that a higher EAp53 score (more severe impact) correlated with worse overall survival. Using statistical methods, a cut-off EAp53 score of 52 predicted for worse OS. Based on this, we classified the patients into 2 risk-groups: 17 (6%) low-EA-MDS with a significantly longer OS (median, 48 months), and 253 (94%) high-EA-MDS with a median OS of 10 months (p=0.01) (Fig. 1A). Interestingly, there was no difference in the distribution of TP53 VAF or R-IPSS scores between this 2 risk-groups. Further analysis revealed that the EAp53 risk group independently predicted OS along with R-IPSS risk score and serum albumin, while other parameters including TP53 VAF and the number of TP53 mutations did not.
Although small, the low-EA-MDS patients formed an important subset with a favorable outcome within TP53 mutated MDS. Upon further investigation, low-EA-MDS patients showed distinctive clinicopathologic features such as lower frequencies of CK, MK, and multi-allelic TP53 alterations suggesting that the type of mutation dictates the degree of karyotypic and genomic complexity. Low-EA-MDS patients often had additional somatic mutations, involving NRAS and RUNX1, that potentially modified the outcome. By immunohistochemistry, low-EA-MDS showed lower TP53 protein expression compared to high-EA-MDS, implying that these mutations did not completely abrogate TP53 function. Interestingly, the 2 risk-groups did not show any significant differences in the distribution of median TP53 VAF, distribution of IPSS-R, or treatment characteristics. By sequential NGS profiling, low-EA-MDS and high-EA-MDS showed similar mutation clearance rates at remission. At progression, in contrast to high-EA-MDS, low-EA-MDS patients acquired additional gene mutations in RAS-pathway genes. These distinctive clinical, cytogenetic, and mutation characteristics provide support to the biologic relevance of EAp53 scoring, substantiating that low and high-EAp53 do not reflect different positions on the disease trajectory.
Another observation was that some patients with similar EAp53 scores had different survival. We hypothesized that this variability may be due to the different locations of these mutations within the 3D protein structure. To explore this, we performed 3D protein structural analysis. TP53 variants with poor OS <10 months clustered in 2 locations: a large cluster interfacing the DNA-binding site and a small cluster formed by residues V157, Y220, L257, and E258, showing that structure location further stratifies the outcome (Fig. 1B).
Finally, we confirmed the prognostic cut-off score of 52 for EAp53 using an independent validation cohort. When compared to other independent computational methods (CADD, REVEL etc), EAp53 better segregated the prognostic subgroups.
How are these findings relevant in the context of upcoming novel mutation type-specific therapies?
Low-EAp53 mutants may benefit from approaches that utilize residual TP53 function while small molecules to restore TP53 function, such as Eprenetapopt (APR-246), may be beneficial for high-EAp53 mutants. Further validation in multi-center cohorts within the context of novel therapies is warranted.
Take-home points:
This is the first study to show the independent prognostic value of the EAp53 score, thereby expanding the previously established genomic attributes impacting the outcomes of TP53-mutated MDS. Unlike VAF and karyotype, EAp53 score is a mutation-dependent, stable predictive biomarker for baseline risk-stratification, not influenced by time or therapy, and can identify prognostic subsets and guide treatment.
To read the full paper, click the link: Full article on Blood Cancer Journal website
Follow the Topic
-
Blood Cancer Journal

This journal seeks to publish articles of the highest quality related to hematologic malignancies and related disorders.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in