Spatial Proteomics Offer Novel Insights into Rare Lymphomas with Sparse Tumor B-Cells
Published in Cancer
NLPHL is a rare lymphoma that is characterized by sparse tumor B-cells, a rich inflammatory tumor microenvironment (TME), and a favorable prognosis. Patients with NLPHL, however, can have multiple recurrences or undergo progression to a large B-cell lymphoma (THRLBCL), which is associated with an aggressive clinical course. NLPHL and THRLBCL are part of a biologic spectrum that share many similarities in immunophenotype, gene expression, and mutational profiles. Previousely, we showed that variant growth patterns of NLPHL is associated with less favorable disease and increased recurrences and/or progression to THRLBCL. Reproducible criteria to separate variant patterns of NLPHL, particularly pattern E, from THRLBCL is still undefined.
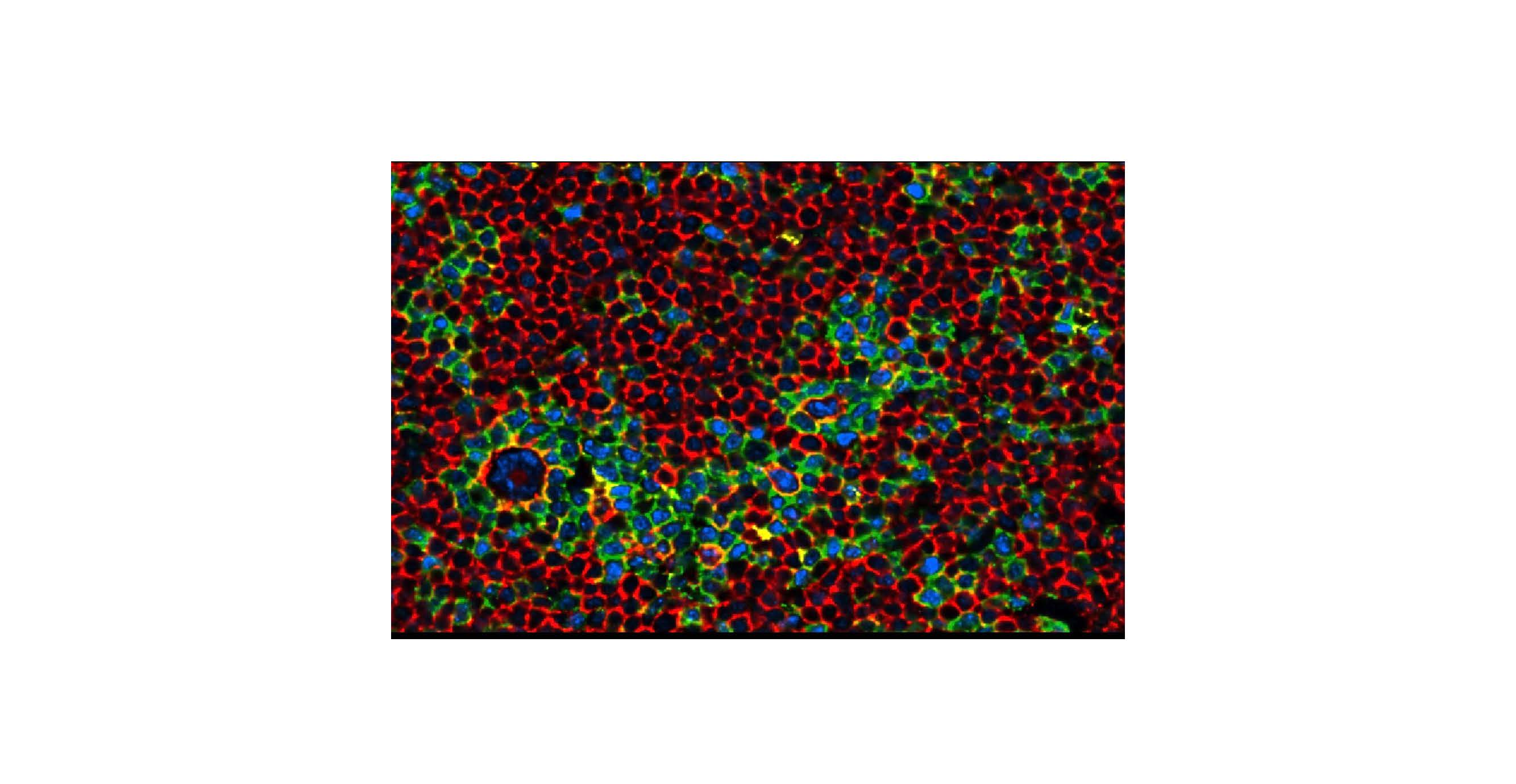
We used co-detection by indexing [CODEX® (rebranded as PhenoCycler™ from Akoya Biosciences)] with a carefully curated, DNA-barcoded 21-antibody panel (CD20, PAX5, CD19, CD22, IGD, CD21, BCL6, MUM1, CD3, CD4, CD8, ICOS, PD1, PDL1, FOXP3, CD69, LAG3, IDO1, MPO, CD14, CD68) that could simultaneouly interrogate tumor B-cells and immune cell subsets. For staining, the 21-antibody panel was applied to 15x15mm representative areas selected by pathologists from 4µm formalin-fixed, paraffin-embedded (FFPE) tissue sections of 20 lymph node excisional biopsies covering the NLPHL growth patterns and THRLBCL. A 96-well plate for the multicycle experiments with different fluorescent oligonucleotides was used with automated image acquisition. Staining parameters including segmentation accuracy were validated by pathologists. For CODEX analysis, four regions of interest (ROIs) were selected per case to ensure that NLPHL patterns and THRLBCL were represented. The selected ROIs in all cases had between 10431-65884, mean+/-SD of 33965.7 +/- 14331.6 number of cells per ROI. Clustering was performed in R using the Seurat package for unsupervised clustering. Cell populations were broadly grouped by phenotype and by activation status and verified at each analysis step by pathologists. QuPath (https://qupath.github.io/) was used to quantify nuclear size and further study macrophage/monocyte markers PU.1, CD163, and CD14 in independent cohorts of 15 NLPHL pattern E and 14 THRLBCL cases.
By deeply profiling the cellular composition and spatial organization of NLPHL and THRLBCL, our findings showed that both lymphomas contain distinct cell compositions and spatial organization. These findings corroborate known interactions as well as identify novel associations regarding cell type abundance, functional status, and spatial distribution of specific cell populations at the single-cell level in tissue samples. We found that tumor B-cell size and content increased gradually from typical to variant NLPHL and was highest in THRLBCL, whereas the trend for TME B-cells was the reverse. This observation is similar to previous studies using 3-dimentional confocal microscopy. The ordered increase in tumor B-cell size from typical to variant NLPHL and THRLBCL was also found to align with the presence or absence of nodularity. These findings raise the possibility that lymphoma progression and dissemination may occur more frequently in less confined architectural configurations, although this hypothesis will require additional study.
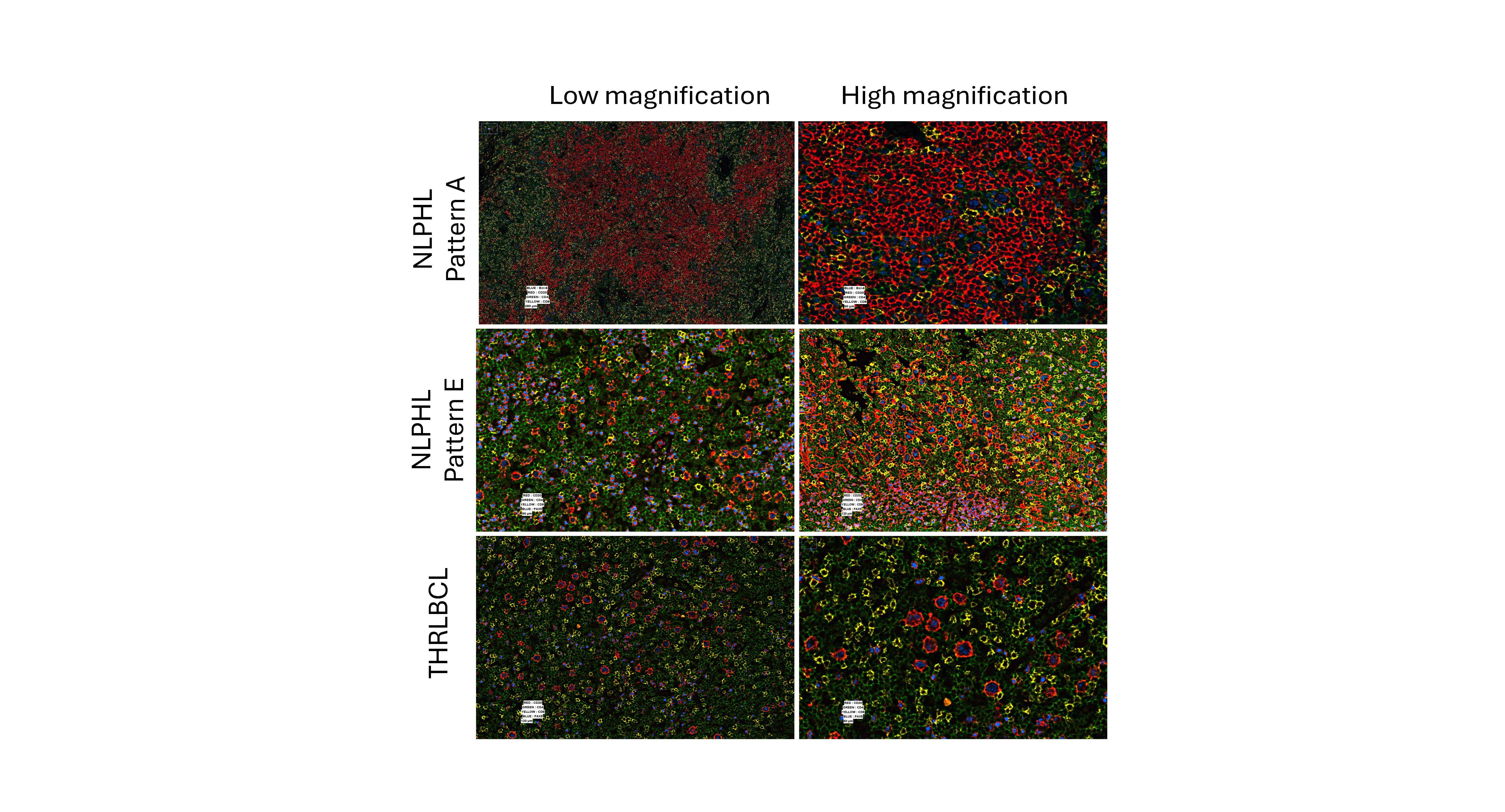
Our results further show that there are marked differences among T-cell and macrophage subsets and spatial configuration among NLPHL patterns and THRLBCL. In the NLPHL TME, there were abundant helper T-cells (CD3/CD4), activated helper T-cells CD3/CD4/LAG3/CD69), and T-follicular helper cells (CD3/CD4/PD1/BCL6), whereas the THRLBCL TME had increased regulatory T-cells (T-regs, CD3/CD4/FOXP3), cytotoxic T-cells (CD3/CD8), and activated cytotoxic T-cell (CD3/CD8/LAG3/CD69). CD4/CD8 double-positive T-cells were present in all NLPHL but not in the majority of THRLBCL cases, and were found to be spatially distant from the tumor B-cells and the T-follicular helper rosettes typically seen in NLPHL. The most striking difference was seen in macrophage/monocyte content in distinguishing NLPHL pattern E from THRLBCL, which we further validated by immunohistochemistry for CD163, PU.1 and CD14 in independent cohorts of cases. These profiles provide a seminal description of the composition and spatial configuration of TH, TFH, Tregs, and their activated counterparts as well as macrophage/monocyte subsets in NLPHL and THRLBCL. Differential annotations were especially aided by the use of LAG3 and CD69 to define activation status among T-cell subsets together with subset-specific markers of other lineages. The differences in T-cell and macrophage subsets are particularly provocative for future work in the derivation of clinical algorithms for tissue diagnosis, particularly in the difficult diagnostic boundary between NLPHL pattern E and THRLBCL.
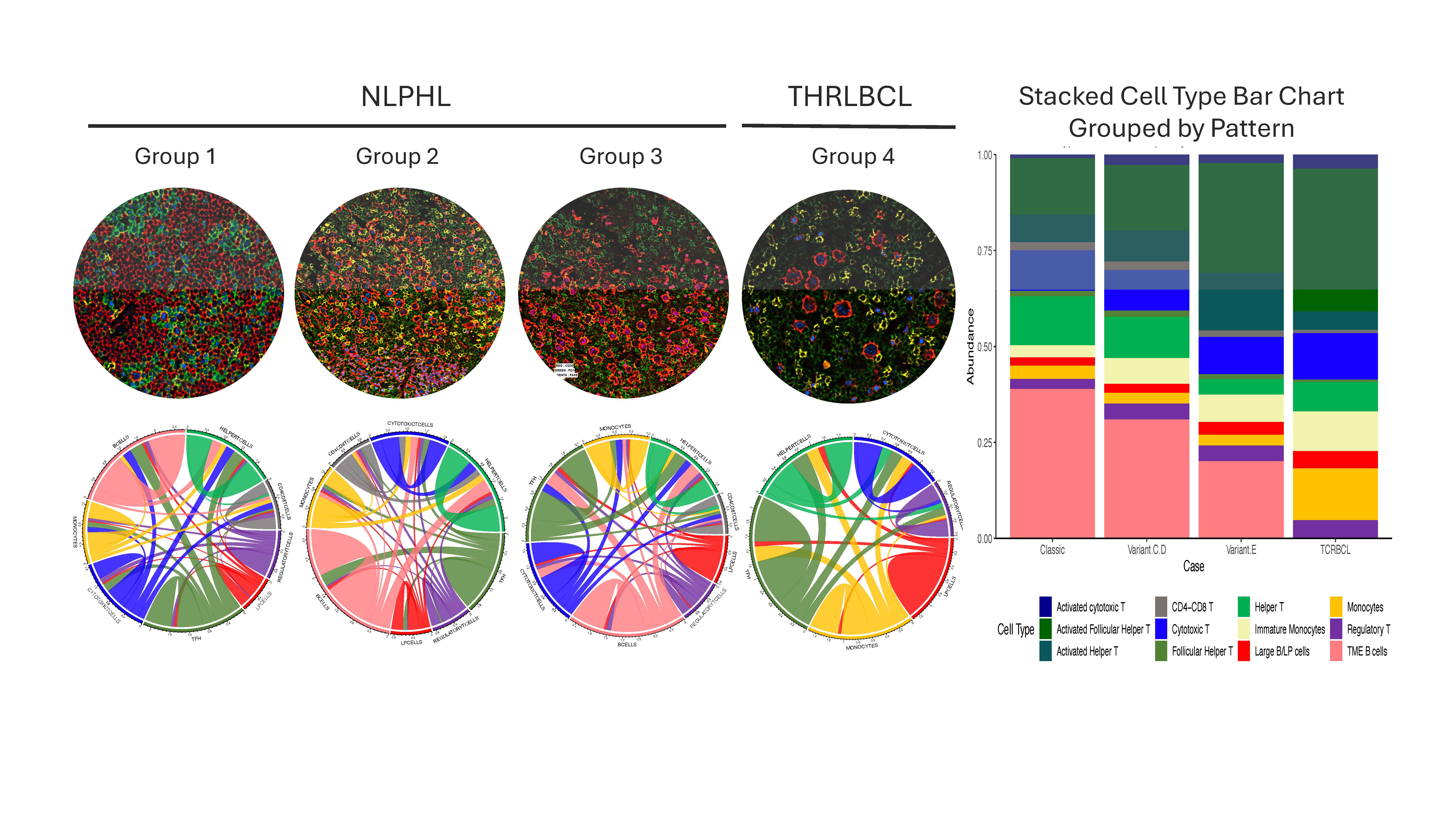
Overall, the use of whole tissue sections with careful annotation by pathologists provided novel findings regarding cell type abundance, functional status, and spatial configuration of NLPHL and THRLBCL at the single-cell level. These findings broadly validate current classification approaches in these lymphomas and provide substantive groundwork for further investigation. Larger retrospective as well as prospective cohorts of clinically well-annotated NLPHL and THRLBCL cases will be necessary to fully decipher the TME and to confirm and validate the diagnostic utility and clinical implications of these findings. In addition, our investigative approach provides a framework to address how spatial proteomics can be a significant tool to define tumor-TME niches and to offer diagnostic precision in cancer biology.
Follow the Topic
-
Blood Cancer Journal

This journal seeks to publish articles of the highest quality related to hematologic malignancies and related disorders.


Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in