Automated design of synthetic microbial communities
Published in Bioengineering & Biotechnology

Microbial communities are observed throughout nature. They consist of multiple species exhibiting genetic and phenotypic diversity. The combination of multiple species in a shared environment can yield emergent properties that could not be demonstrated by a single independent species. Communities can be seen to separate tasks between subpopulations (division of labour) which can increase productivity through metabolic specialisation of species. By making full use of the resources in the environment and collaborative defence mechanisms, communities also possess a resistance to pathogenic invasion. The emergent properties of microbial communities make them attractive to biotechnology applications, where specialisation of species can be used to improve bioprocess productivity. Additionally, our increasing understanding of the role microbiomes have in disease (particularly gastrointestinal), makes manipulating microbial communities a target for therapeutics.
Microbial communities do not form by simply mixing multiple species together. Differences in species growth rate and fitness will normally result in the most competitive strain occupying mutual limiting resources and driving the remaining species to extinction. Communities have a better chance of forming when the species involved communicate with one another to coordinate growth and consumption of resources, enabling their coexistence and prevent dominance of individual species. The types of interaction in a community will influence the type of population dynamics we see. Stability, oscillations and even chaos have all been observed in microbial communities. Understanding the interactions that enable the formation of microbial communities and how they can inform the population dynamics is an extremely important step required to improve our ability to build microbial communities for biotechnology applications, and manipulate existing microbiomes to treat disease.
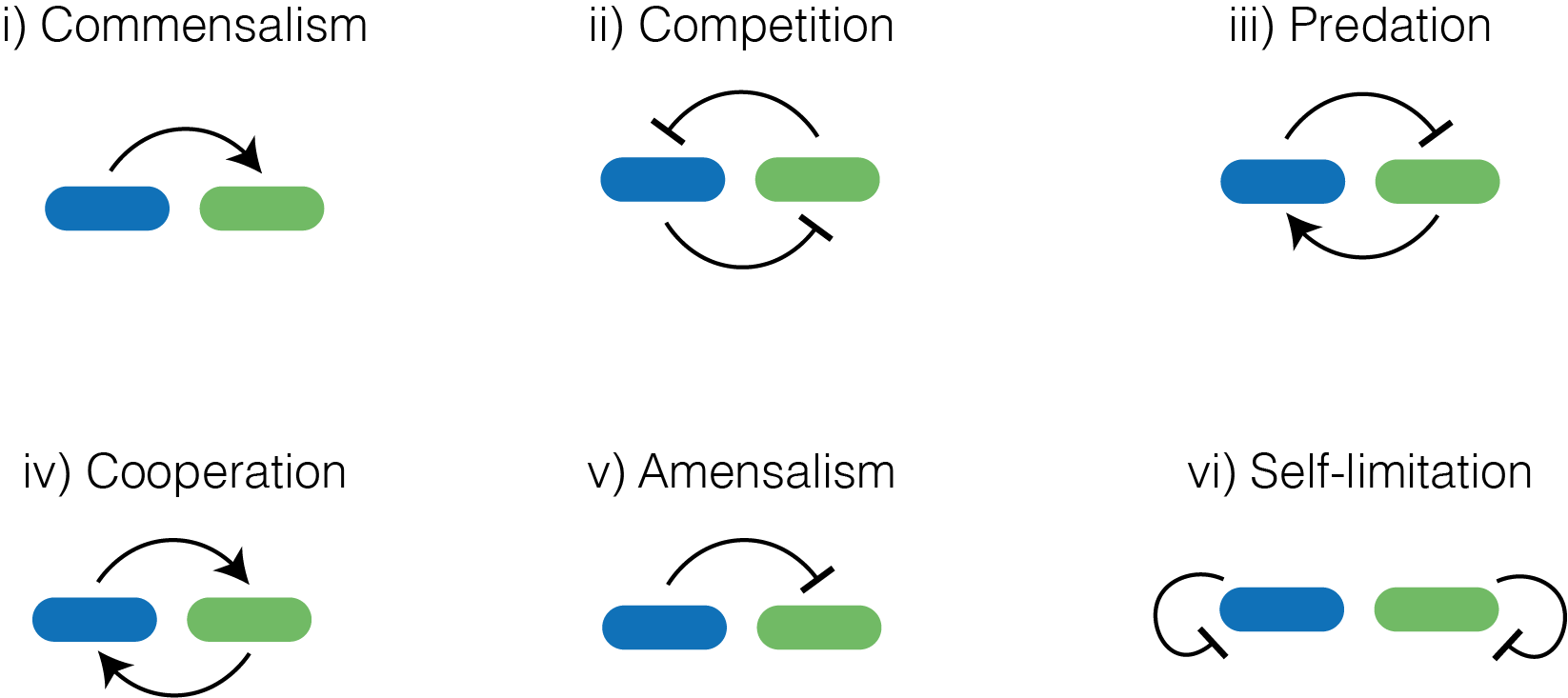
Figure 1 Fundamental interactions that we can engineer by combining quorum sensing and bacteriocins
In this paper we investigate interactions that can be built using quorum sensing systems and bacteriocins. Quorum sensing systems are a form of cell-cell communication that can be found in microbes, and can be used to regulate gene expression. Bacteriocins are a type of antimicrobial peptide that can be produced by bacteria. They have a negative effect on the growth of sensitive strains. By combining quorum sensing and bacteriocins, we possess the tools to engineer communication and interactions between strains that can engineer fundamental ecological interactions (Figure 1).
We used methods from Bayesian statistics to identify the combinations of quorum sensing systems, bacteriocins and strain sensitivities that show the highest probability of producing stable communities of two and three strains. We were able to derive the fundamental interactions that are most commonly associated with stable communities, and demonstrate parameter regions where stability has the highest probability of occurring. These results feed into our future lab work, informing the design of microbial community topologies, and the genetic parts we use to build them.
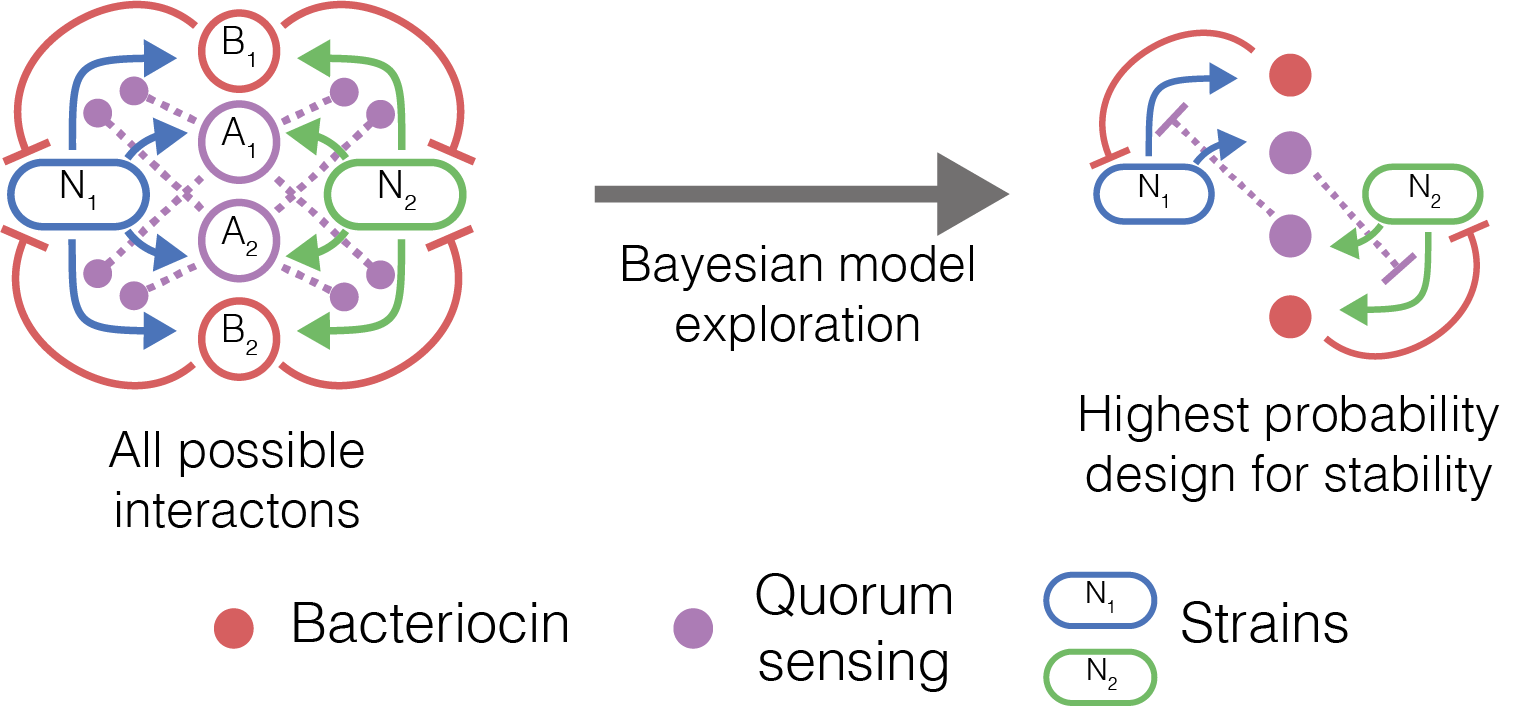
Figure 2 Overview of our work in two species community, showing all the possible interactions we can engineer using two quorum sensing systems and two bacteriocins (left). The top performing model (right) we identified consists of cross protection mutualistic relationships.
This work reinforced the need for computational tools to guide us in designing microbial communities. When faced with a large number of possible designs, deciding design which has the highest probability of success is extremely hard when using intuition alone. Methods such as these help us make data driven decisions in the lab, improving our opportunities for success.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in