Beggiatoa's lipoxygenase: does a close relative of Thiomargarita magnifica carry a biochemical secret?
Published in Ecology & Evolution, Microbiology, and Cell & Molecular Biology

Lipoxygenase: a marker of multicellularity?
Lipoxygenase is familiar to any biomedical specialist as a key enzyme in the production of leukotrienes and lipoxins — lipid substances involved in the regulation of inflammation. But its role goes far beyond — it is an universal cell-to-cell signalling enzyme across multicellular eukaryotes: stress-related jasmonate pathway in plants, quorum sensing in fungi and even pheromone biosynthesis in brown and diatom algae! In all these groups, lipoxygenase starts the synthesis of oxylipins — oxygenated polyunsaturated fatty acid derivatives — which, in turn, act as signalling compounds. Given the high probability that such different groups of eukaryotes evolved cell-to-cell signalling independently, this similarity raises a question: why do they use the same biochemical solution to arrange it?
We don’t have the full answer yet. But, in a paper published in 2020, my co-authors and I presented a bioinformatic study suggesting that lipoxygenases could be associated with multicellularity both in bacteria and eukaryotes — even in the most primitive forms [1]. We found both statistical and evolutionary correspondences between lipoxygenase and multicellularity. This paper was earlier presented here in a blog post: “Bacterial oxylipins: a key to multicellularity and to combating antimicrobial resistance?”
We have long accustomed to the concept of multicellularity as something big, complex, having organs, and clearly visible to the naked eye — but this is the tip of the multicellularity iceberg. As I have mentioned earlier in a blog post “What is to be multicellular? A question from a novel cave bacterium”, some multicellular organisms, like slime moulds (Fig. 1) and myxobacteria, construct their own temporary bodies from different free-living cells (this type of multicellularity is referred to as aggregative). These bodies are typically small and simple enough to be omitted in fallen leaves or in rotten straw. Other multicellular organisms are clonal, as we are — this means that their bodies descend from one cell each. But some of them form thin filaments one cell thin (like cyanobacteria, Fig. 2), and this is the reason why some of us didn’t conceive the idea that they are actually multicellular just like you or at least a flower on your windowsill. There are a lot of multicellular organisms with a much simpler multicellularity that you could imagine.
We found that lipoxygenase is associated even with such simple types of multicellularity across both bacteria and eukaryotes. It seems that, once multicellularity emerges somewhere, lipoxygenase is borrowed by a horizontal gene transfer. We don’t know yet what is the cause of its indispensability — but, if our assumptions are true, lipoxygenase could be the most universal biochemical marker of multicellularity we know.
The statistical and phylogenetic picture that impressed us so much was created mainly by cyanobacteria, myxobacteria, dictyostelid slime moulds and complex multicellular eukaryotes. These taxa showed both the high lipoxygenase prevalence and phylogenetic pattern of lipoxygenase acquisition simultaneously with the rise of multicellular forms. But there was one more multicellular bacterial taxon in our sample of bacteria for which lipoxygenase sequences were available in NCBI and UniProt databases. This was Beggiatoa — a genus of giant sulfur bacteria.
Beggiatoa: sulfur monsters
Their multicellular forms are similar to cyanobacteria — Beggiatoa live as long filaments consisting of individual cells. Inside these cells, yellow light-refracting granules are visible — they are nothing else but the globules of elemental sulfur (Fig. 3, Video below). This is a crucial biochemical intermediate deriving in the process of oxidizing sulfide to sulfate which gives the energy to this bacterium. If you oxidize organic substances contained in your hamburger and earn your ATP, Beggiatoa oxidizes sulfide instead. This way of thriving is called chemolithoautotrophy (while you are chemoorganoheterotrophic).
This way of life forces Beggiatoa, like other sulfur bacteria, live in habitats where sulfides are always available (like sulfur caves or sulfide-rich lakes, Fig. 4). But, to oxidize sulfur, Beggiatoa needs oxygen. It lives at the thin interface of sulfide-rich sediment and oxygen-rich water, and its long filaments bridge together the sulfide pool and oxygen pool. Multicellularity for Beggiatoa is a chance to form long filaments and thus keep a foot in both worlds.

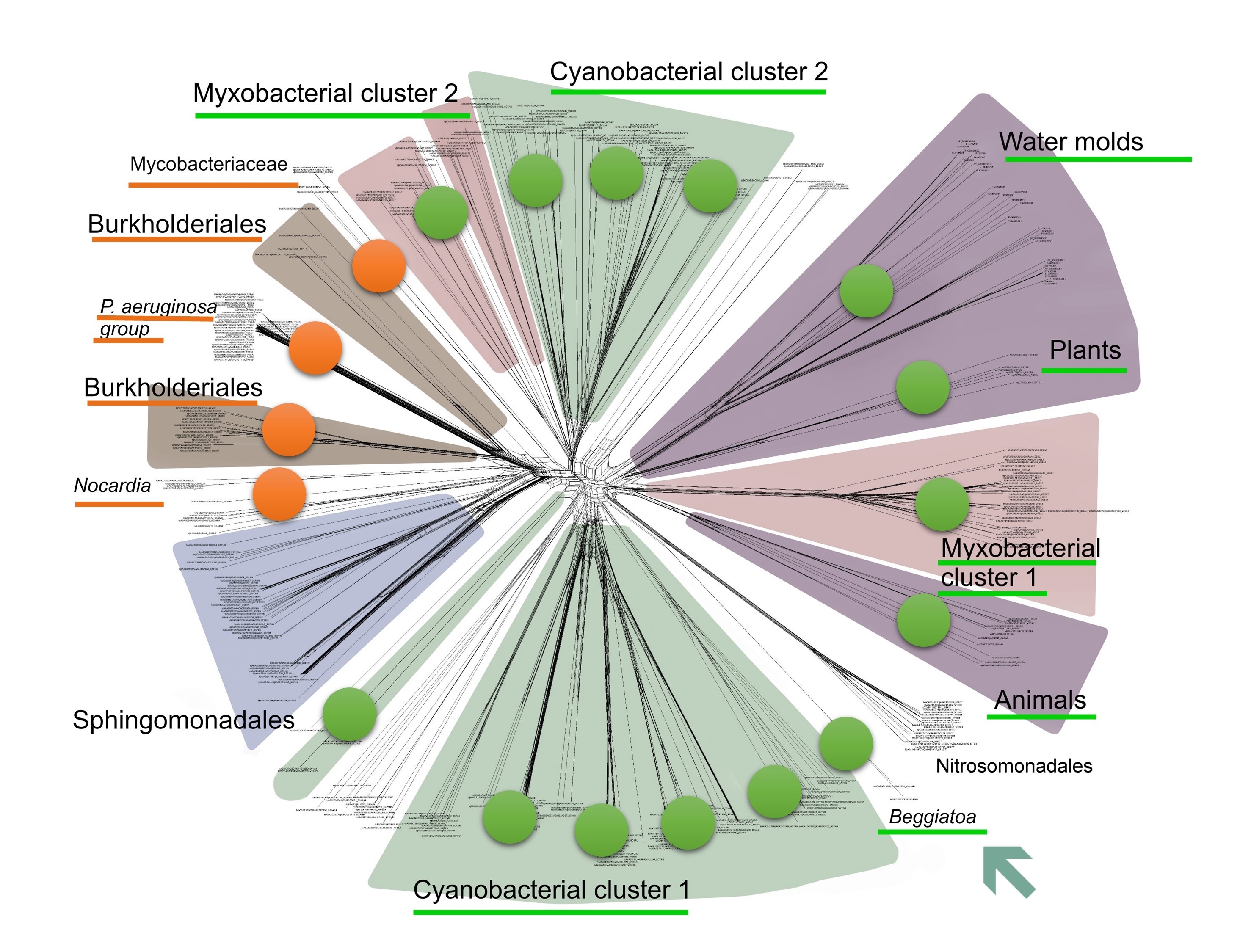
In our earlier work, we did spot the cooccurrence of multicellularity and lipoxygenases in Beggiatoa, but have omitted to draw any conclusions regarding this bacterial genus. Beggiatoa comprises only 2 species (Beggiatoa leptomitoformis and Beggiatoa alba) within the order Thiotrichales. Their lipoxygenases formed a thin and long branch in our phylogenetic models which disallowed any evolutionary assumptions (Fig. 5). Moreover, the lipoxygenase occurrence was low even within the order Thiotrichales — in contrast to other multicellular bacteria for which this rate was high (Fig. 6). In non-mathematical "humanities-like" terms, many Thiotrichales bacteria form long filaments, and many of them are multicellular, but only two species of Beggiatoa have putative lipoxygenases. This was not enough for a bioinformatic study. Puzzle pieces didn't fit together.
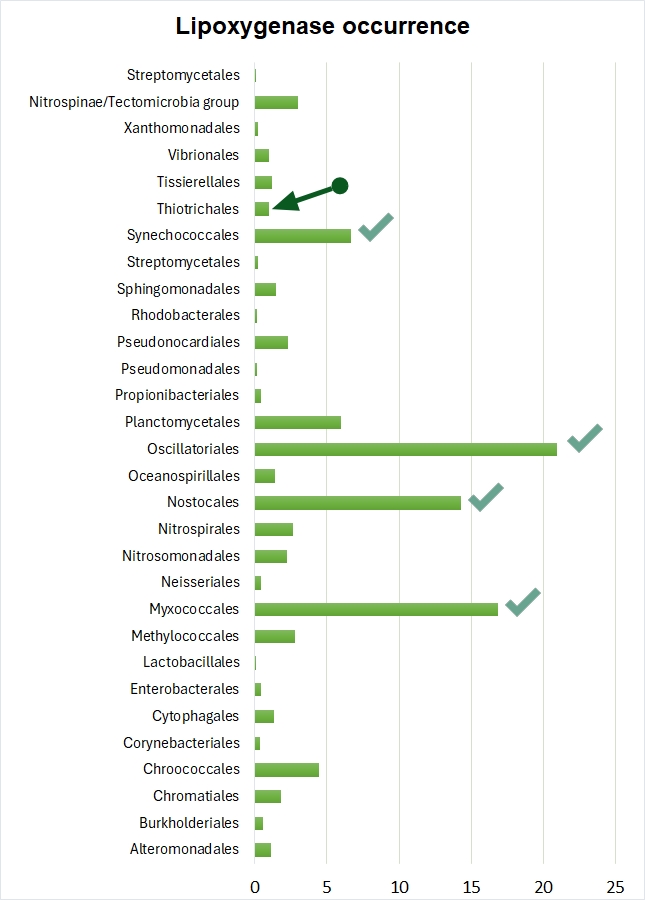
Thiomargarita magnifica: the missing piece of the puzzle
One day, I traveled by a commuter train from work to home and read a recent issue of New Scientist which had just arrived to me by subscription. The article on the discovery of Thiomargarita magnifica (Fig. 7) captured my attention. I have previously met the news of a novel giant bacterium out of the corner of my eye, but this was the time I looked deep.
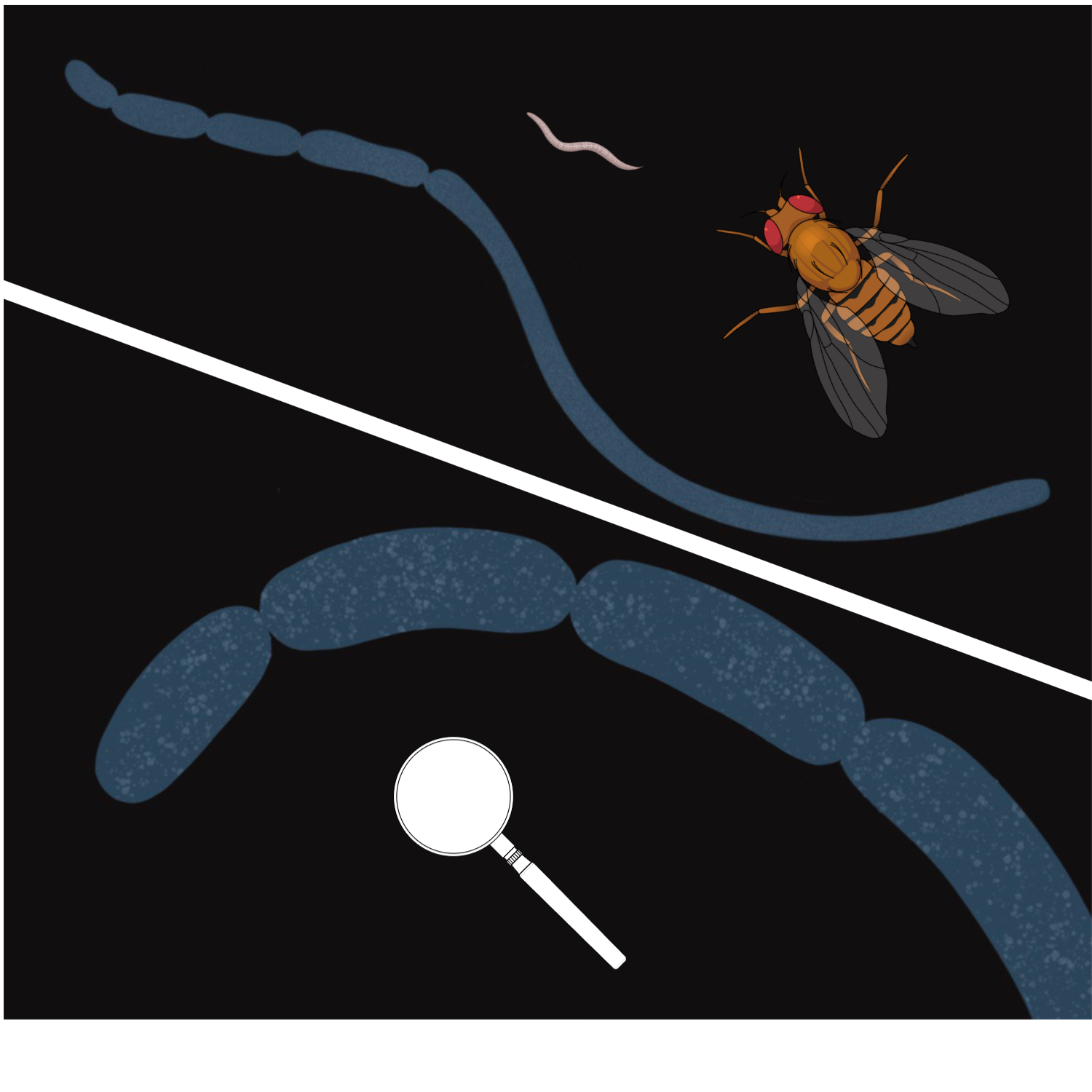
The newly discovered bacterium — a giant of the bacterial world which is larger than Drosophila fruit fly — was the close relative of Beggiatoa [2]. It came to be a sulfur bacterium with similar metabolic demands. These representatives of the same bacterial order Thiotrichales need to bridge two spatially separate pools — the pool of oxidant and the pool of sulfide — just like Beggiatoa and other giant sulfur bacteria.
I felt thunderstriken, in the finest sense of the world. It was a stroke of insight. Thiomargarita magnifica appeared as a missing piece of the puzzle. Being the closest relative of Beggiatoa and Thioploca, it also reached the incredible complexity by increasing its size — but did this without multicellularity. Its giant filament is a single cell, which is highly unusual way to be large. Cells have fundamental constraints for their size — their energy is generated by ATP synthases which can be located only on membranes. The number of available ATP synthases, in turn, generally depends on the total size of membranes. The bigger the cell, the smaller the surface-to-volume ratio — and the less energy is available for the cell metabolic machinery. But Thiomargarita magnifica has increased the intracellular complexity almost to the eukaryotic level — its ATP-synthases are located on the cytoplasmic membrane bubbles called pepins, which also contain the DNA. This drastically increases the ATP-generating surface.
Two close relatives, Thiomargarita and Beggiatoa live in similar conditions and are exposed to the same selection pressure urging to be larger — but solve the problem in different ways. This means that Thioploca and Beggiatoa also could find the way to bridging the gap independently. And this could be the explanation why only Beggiatoa acquired lipoxygenases.
New discussion article
Thioploca relies more on its motility — it can more actively glide up and down to refill the metabolic storages, then "hold the breath" and perform necessary biochemical processes. Beggiatoa are less motile, and their only strategy to keep the enough length is multicellularity. Maybe, it was the reason why this bacterium acquired lipoxygenases while other bacteria didn't (Fig. 8).
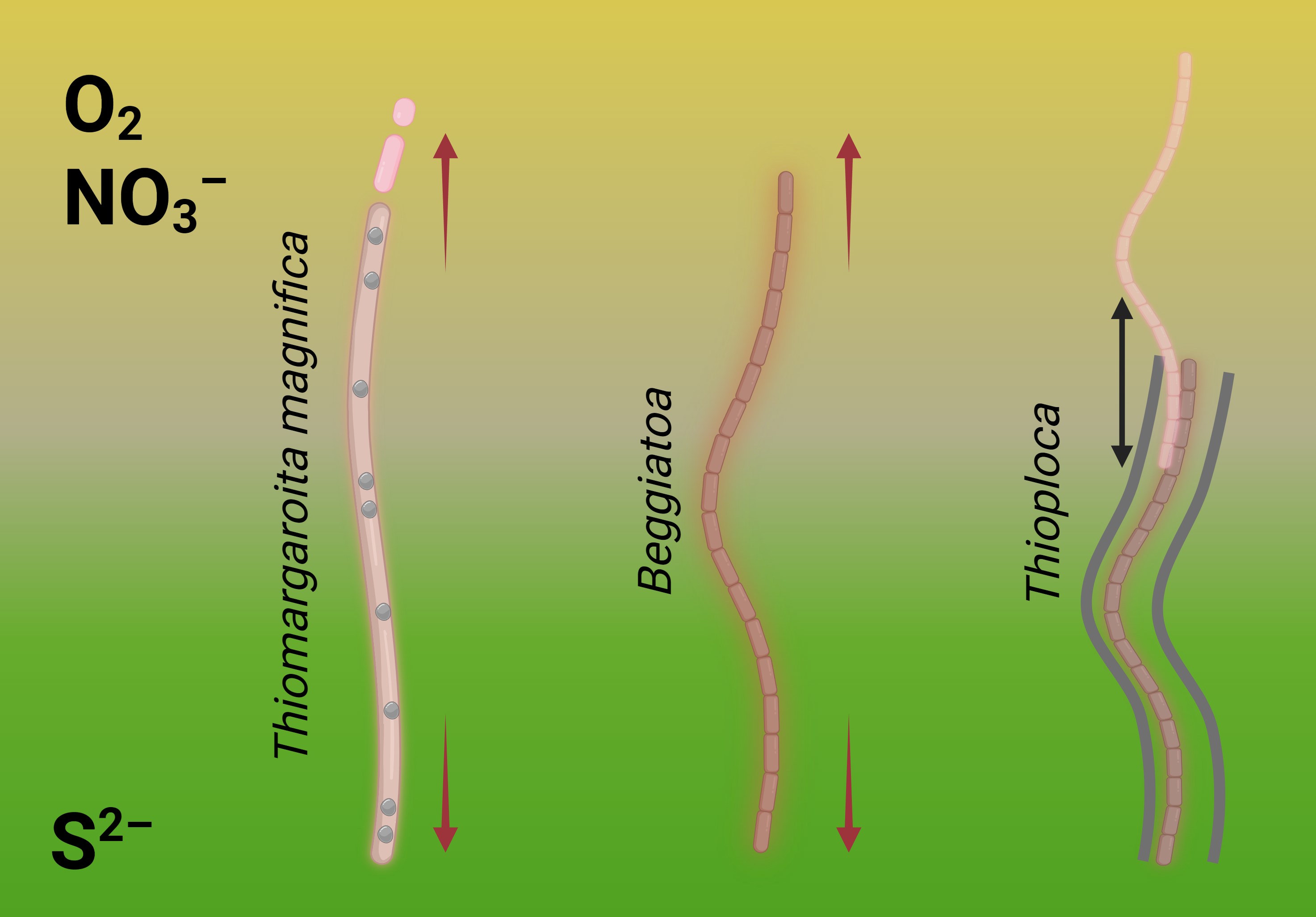
This hypothesis is only one of different biochemically plausible explanations — but it outlines the main idea that the ways to complexity may be very different within Thiotrichales. This has been proven by Thiomargarita magnifica. Beggiatoa's lipoxygenase is likely onу of such peculiar ways, and Beggiatoa may be justified — they probably use lipoxygenases to maintain their multicellularity despite the low statistical rates.
I reflected this newly discovered evidence in a new short discussion article in Biochemistry (Moscow) [3]. There, I summarized the ideas explained above. I hope this short article will really spark a discussion — the more so, multicellularity-related lipoxygenase in a sulfur bacterium raise a lot of questions. Our knowledge about oxylipin signalling in bacteria is very scarce, but at least, we know some biochemical facts about lipoxygenases in cyanobacteria and myxobacteria — other multicellular bacteria. But for Beggiatoa, we know nothing about their lipoxygenases — and they could come to be a new biochemical planet discovered with the point of our pen.
Acknowledgements
I say a lot of thanks to biological artist Anastasiia Samoukina, who created artistic pictures above. She is a regular illustrator of my lipoxygenase evolution project, and it would not be such beautiful without her invaluable help.
YouTube video by Journey to the Microcosmos.
References
- Kurakin G.F., Samoukina A.M., Potapova N.A. (2020) Bacterial and Protozoan Lipoxygenases Could be Involved in Cell-to-Cell Signaling and Immune Response Suppression. Biochemistry (Moscow), 85, 1048–1063. https://doi.org/10.1134/S0006297920090059
- Volland J.-M., Gonzalez-Rizzo S., Gros O., et al. (2022) A centimeter-long bacterium with DNA contained in metabolically active, membrane-bound organelles. Science, 376, 1453-1458. https://doi.org/10.1126/science.abb3634
- Kurakin, G. (2023) Lipoxygenase in a Giant Sulfur Bacterium: An Evolutionary Solution for Size and Complexity? Biochemistry (Moscow), 88, 842–845. https://doi.org/10.1134/S0006297923060111
UPD from 11.12.2023: Figures and captions updated.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in