Cultured meat with enriched organoleptic properties by regulating cell differentiation
Published in Bioengineering & Biotechnology, Materials, and Agricultural & Food Science

Research Background
Cultured meat is arising attention as a sustainable meat which can save the natural resources consumption and pollutant emission involved in breeding livestocks. The main aim of cultured meat is on mimicking the physical and sensorial properties of conventional meat. To implement this, scaffold can play a critical role by regulating cell growth to determine the biolgical characteristics of cultured meat. However, it is veiled that how sensorial and nutritional characteristics of cultured meat are affected by cellular behavior.
Research Strategy
Important factors which determine the quality of meat such as texture, flavor, and nutrition values are derived from the biological and chemical properties of muscle and fat tissues. Specifically, dimensions and amount of muscle proteins in muscle tissue affect the texture and nutrition facts of meat whereas lipid amount in fat tissues affect the flavor of meat. Because these molecules are synthesized by cells comprising the tissues, we hypothesized that regulating the synthesis of these cell-derived molecules would also affect the organoleptic properties of cultured meat. Therefore, control of cell differentiation was performed using scaffold engineering technique. Then, different synthesis of muscle proteins and lipids were confirmed depending on the cell differentiation degree. Finally, the effect of these differences on the organoleptic properties of cultured meat was deeply investigated.
Results
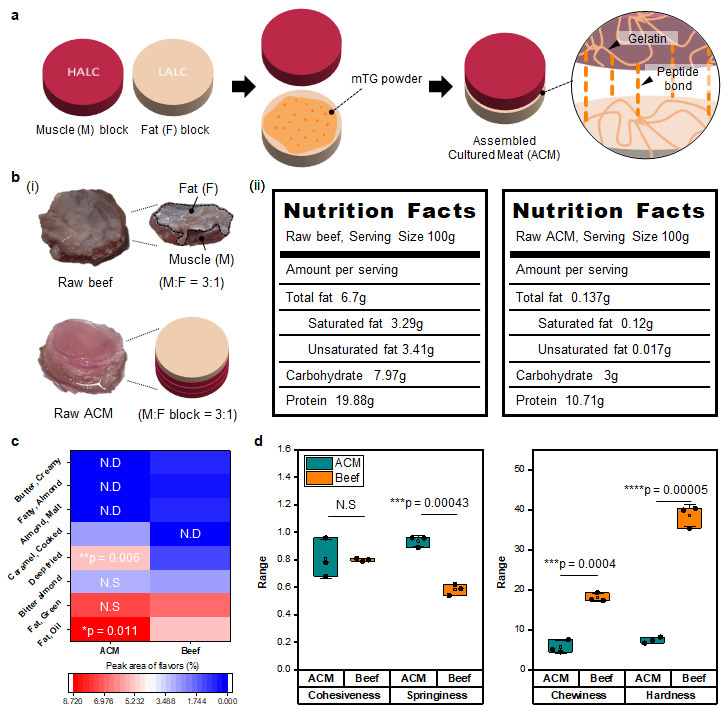
To regulate cell differentiation, we fabricated gelatin/alginate double network scaffolds with different stiffnesses. Crosslinking degree of alginate network was controlled by calcium ion concentration to vary mechanical properties between the scaffolds. As a result, we obtained two types of scaffolds: 1. Scaffold with muscle-like stiffness (Young’ s modulus ~ 11 kPa), 2. Scaffold with fat-like stiffness (Young’s modulus ~ 3 kPa). Then, these two scaffolds were applied to bovine myoblasts and myogenic differentiation was induced. The amount of myosin heavy chain, a representative muscle protein derived from myogenic differentiation, was dependent on the degree of myogenic differentiation. Importantly, this protein amount difference signicantly affected the texture and flavor of cultured meat. Because muscle proteins aggregate and stiffen upon heating, they are known to determine texture of conventional meat. In our system also, the amount of muscle proteins was confirmed to affect the texture of cultured meat. In addition, proteins reacted with sugars upon cooking temperature and generated various Maillard reaction products when the cultured meat was cooked. Moreover, adipogenic differentiation was regulated depending on the scaffold’s stiffness resulting different amount of cell-derived lipids. When cultured meat contained different amount of lipids, fatty flavor intensity was varied. From these results, we confirmed that regulating cell differentiation is critical to control the organoleptic properties of cultured meat. Muscle blocks and fat blocks which contain highly differentiated muscle and fat cells respectively were finally assembled together to fabricate the cultured meat with marbled structure. Using our strategy, we confirmed that the organoleptic properties and nutritional value of the assembled cultured meat became similar to those of conventional beef.
Conclusion
We reported an innovative strategy to produce high-quality cultured meat which mimics the sensorial properties of conventional meat. This study revealed that the degree of cell differentiation significantly affects the various organoleptic factors of cultured meat. We believe that our study is cornerstone research which can contribute to future research on cultured meat development.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in
This is an outstanding study. Congratulations on this important contribution to cultured meat research. I’m a biologist from Peru, and I’m currently exploring the application of these approaches using native species such as the guinea pig (Cavia porcellus), which is culturally significant in Andean regions. I would love to connect and learn more about your methodology.