Decoding the Hidden Microbiome Behind Triple-Negative Breast Cancer
Published in Cancer, Microbiology, and General & Internal Medicine
Triple-negative breast cancer (TNBC) is one of the most aggressive forms of breast cancer. Although it accounts for only about 10–15% of all breast cancer cases, it is responsible for nearly 40% of breast cancer-related deaths. Unlike other subtypes, TNBC lacks hormone receptors such as estrogen, progesterone, and HER2, which means that targeted therapies commonly used in other breast cancers are ineffective. This makes TNBC harder to treat, more prone to relapse, and more likely to spread.
In recent years, scientists have started looking beyond human genetics and tumor biology to understand what drives cancers like TNBC. A surprising area of discovery has been the breast tissue microbiome—the community of bacteria that resides in breast tissue. Once thought to be sterile, breast tissue is now known to host a variety of microbes, some of which may influence cancer development and progression.
Yet, despite growing evidence of microbiome involvement in breast cancer, the microbial ecosystem of TNBC remained largely uncharted. That is where our work comes in.
What We Did
Our team set out to answer a simple but powerful question: Are there microbial players specific to TNBC that could help explain why it is so aggressive?
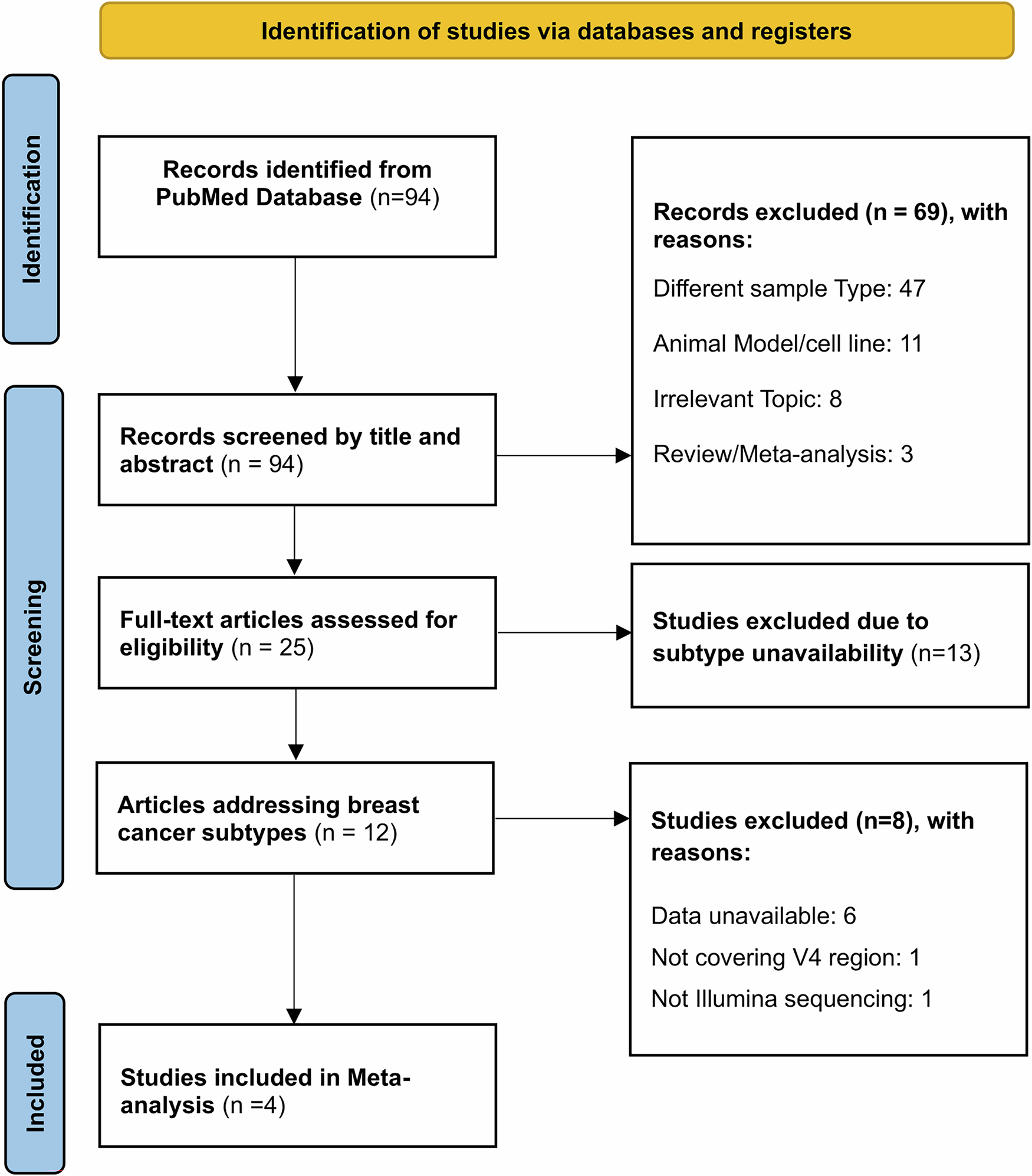
To do this, we conducted the first meta-analysis of the TNBC tissue microbiome. A meta-analysis is important because it combines data from multiple independent studies, increasing both statistical power and reliability. Instead of relying on a single study with a limited sample, we aggregated raw sequencing data from 200 breast cancer tissue samples across four cohorts from the United States, China, and Italy.
This “mega-analysis” approach was particularly important because microbiome studies can differ in methods and sequencing strategies, making results difficult to compare directly. By standardizing preprocessing and applying robust statistical corrections, we ensured that microbial differences we observed were genuine and not artifacts of study design.
We then compared the microbial communities in TNBC tissues against those in non-TNBC breast cancers (such as hormone receptor-positive subtypes). Our analysis involved three layers:
-
Diversity analysis – How many different microbes were present, and how evenly distributed were they?
-
Taxonomic analysis – Which specific bacteria were enriched or depleted in TNBC?
-
Functional analysis – What metabolic and signaling pathways were these microbes associated with?
We also applied machine learning models to see if microbial signatures could reliably distinguish TNBC from other breast cancer types.
What We Found
1. TNBC has lower microbial diversity
Compared to non-TNBC tissues, TNBC samples had reduced microbial diversity. In ecology, diversity often reflects resilience. A less diverse microbial community might indicate a disrupted or imbalanced environment—potentially contributing to tumor aggressiveness.
2. Novel microbial players are enriched in TNBC
We discovered three bacterial species that were consistently and significantly enriched in TNBC tissues:
-
Gemmiger formicilis
-
Anaerobutyricum soehngenii
-
Azospirillum oryzae
These microbes have not been previously linked to TNBC in a systematic way. Two of them, Gemmiger formicilis and A. soehngenii, are butyrate-producing bacteria. Butyrate is a short-chain fatty acid with complex, sometimes contradictory roles in cancer. While high concentrations of butyrate can suppress tumor growth by promoting cell death, low concentrations can actually stimulate cancer cell proliferation and migration. This so-called “butyrate paradox” suggests that the presence of butyrate producers in TNBC may fuel tumor aggressiveness under certain conditions.
The third player, Azospirillum oryzae, is even more intriguing. We found evidence suggesting that this microbe may influence TNBC progression through metabolite signaling and immune modulation. For example, it may generate lysophosphatidylethanolamine (LysoPE), a molecule that promotes cell migration and metastasis in TNBC. This is the first report of A. oryzae being specifically enriched in TNBC tissue.
3. Functional pathways linked to cancer hallmarks
The microbes enriched in TNBC were not just bystanders—they were associated with pathways directly linked to cancer hallmarks. These included:
-
Steroid biosynthesis (affecting hormone-related metabolism)
-
Amino acid and fatty acid metabolism (fueling tumor growth)
-
ABC transporters (known to promote metastasis and drug resistance)
-
Quorum sensing (microbial communication that may enhance invasion and angiogenesis)
Taken together, these findings suggest that TNBC’s unique microbial environment actively contributes to its aggressiveness.
4. Machine learning can predict TNBC from microbial signatures
Finally, our machine learning models demonstrated that microbial data alone could distinguish TNBC from other breast cancer subtypes with up to 81% accuracy. This suggests that microbial signatures may serve as potential biomarkers for diagnosis or prognosis.
Why This Matters
Our study makes three key contributions to cancer research:
-
New biological insights – We identified novel bacterial species enriched in TNBC, shedding light on non-genetic factors that may drive its aggressiveness.
-
Potential biomarkers – The unique microbial signatures we uncovered could serve as diagnostic or prognostic biomarkers, complementing existing clinical tools.
-
Therapeutic possibilities – If microbes and their metabolites play an active role in TNBC progression, targeting them could open the door to microbiome-based therapies. This could range from antibiotics and probiotics to dietary interventions and metabolite inhibitors.
In other words, our work highlights the microbiome not just as a passenger in cancer, but as a potential co-driver—one that we may be able to manipulate for better patient outcomes.
The Road Ahead
While our study is the first comprehensive meta-analysis of the TNBC tissue microbiome, it also raises important questions:
-
Are these microbes causal or merely associated with TNBC?
-
How exactly do butyrate levels and microbial metabolites shape tumor behavior in vivo?
-
Can microbiome-targeted interventions improve TNBC treatment outcomes?
Answering these questions will require experimental validation in cell and animal models, as well as metabolomic profiling to directly measure microbial activity. But the direction is clear: the microbiome deserves a central place in TNBC research.
Conclusion
Triple-negative breast cancer is notoriously aggressive and resistant to current therapies. By mapping its unique microbial landscape, our study uncovers hidden players that may help explain its behavior and point toward new solutions.
We discovered three key microbes—Gemmiger formicilis, Anaerobutyricum soehngenii, and Azospirillum oryzae—alongside cancer-linked microbial pathways and diagnostic potential. These insights mark an important step toward microbiome-informed oncology, where understanding and harnessing microbial ecosystems could ultimately improve outcomes for patients facing one of the toughest forms of breast cancer.
read more here: https://link.springer.com/article/10.1038/s41522-025-00816-5?utm_source=rct_congratemailt&utm_medium=email&utm_campaign=oa_20250909&utm_content=10.1038/s41522-025-00816-5
Follow the Topic
-
npj Biofilms and Microbiomes

The aim of this journal is to serve as a comprehensive platform to promote biofilms and microbiomes research across a wide spectrum of scientific disciplines.
Related Collections
With Collections, you can get published faster and increase your visibility.
Natural bioactives, Gut microbiome, and human metabolism
Publishing Model: Open Access
Deadline: May 20, 2026
Harnessing plant microbiomes to improve performance and mechanistic understanding
Publishing Model: Open Access
Deadline: Jun 01, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in