Mapping the AI Frontier in Drug Interaction Prediction: Building Blocks for Egypt HealthGPA
Published in Cell & Molecular Biology, Computational Sciences, and Pharmacy & Pharmacology
Introduction: Why drug interactions matter
Every year, thousands of patients worldwide suffer harm because of unexpected drug interactions. These can occur between two drugs, between a drug and a disease, or even between drugs and the foods or supplements people consume. Some interactions are mild, but others can be life-threatening—leading to hospitalizations, treatment failures, or even fatalities.
Traditional laboratory and clinical approaches to detect interactions are often slow, costly, and limited in scope. As medicine becomes increasingly personalized, there’s an urgent need for faster, scalable tools that can predict risky interactions before they occur.
This is where artificial intelligence (AI) comes in. With the explosion of biomedical data—from drug databases to electronic health records—AI provides the analytical power to uncover hidden patterns, anticipate interactions, and suggest safer treatment pathways.
Our team at Nile University’s Centre for Informatics Science, as part of the Egypt HealthGPA project, set out to systematically map this emerging field. We wanted to answer a foundational question: how far has AI progressed in predicting the broad spectrum of drug interactions, and where are the gaps that remain?
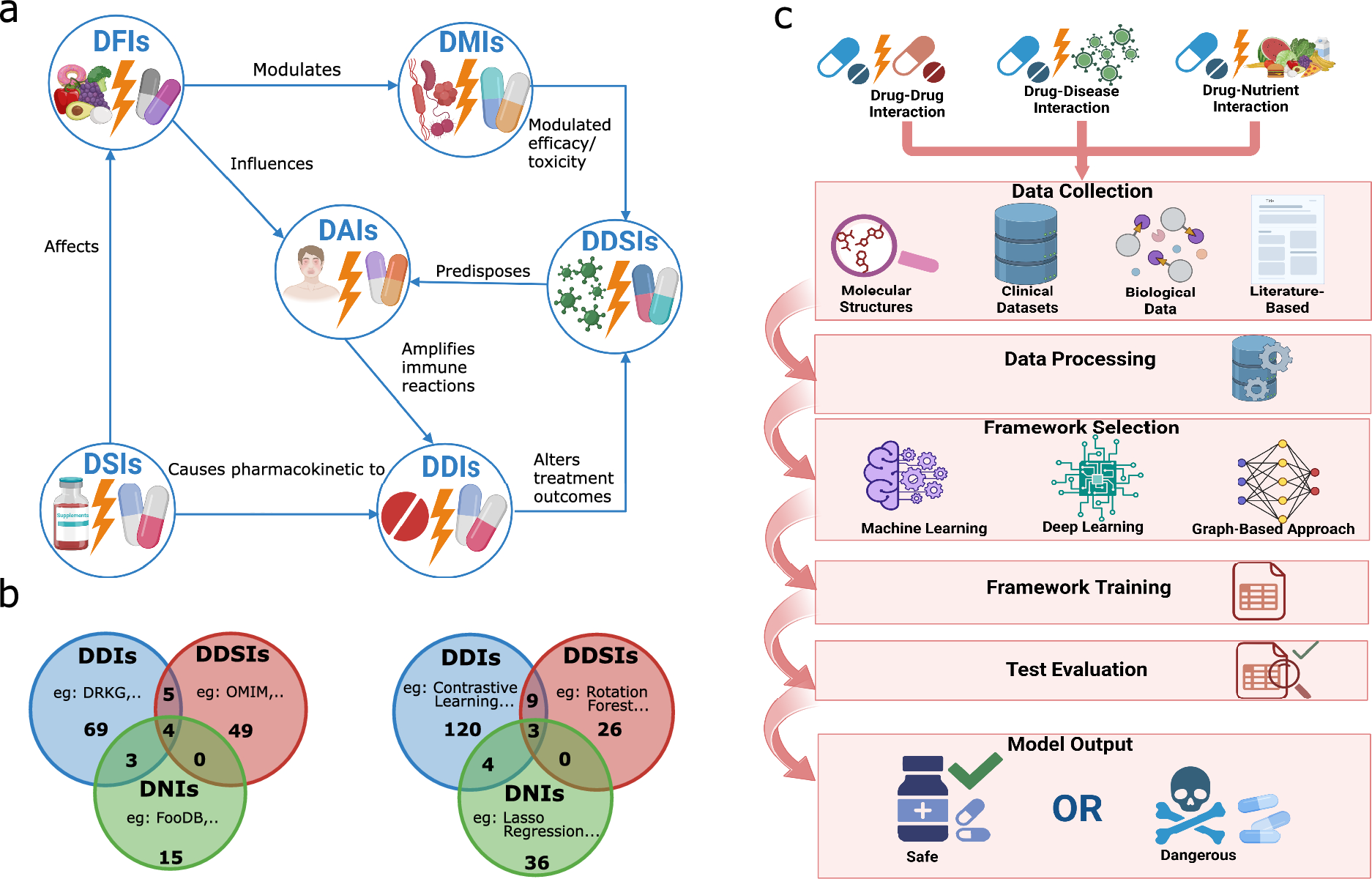
What we did: The first comprehensive AI landscape for drug interactions
We performed a systematic review of 147 studies published between 2018 and 2024 that used AI to predict drug interactions of multiple kinds:
-
Drug–drug interactions (DDIs): when one drug affects another’s absorption, metabolism, or effect.
-
Drug–disease interactions (DDSIs): when a drug worsens or alters the course of an existing condition.
-
Drug–nutrient interactions (DNIs): when foods, supplements, or the gut microbiome interfere with drugs.
-
Drug–allergy interactions (DAIs): when a drug triggers harmful immune responses in predisposed patients.
To ensure rigor, we followed the PRISMA guidelines for systematic reviews, carefully screening thousands of papers and categorizing models, datasets, and outcomes.
Our goal was not only to summarize the state of the art but also to build a taxonomy of methods and resources—a kind of “map of the AI landscape” in drug interaction prediction.
What we found: A field in rapid growth, but with critical blind spots
-
Drug–drug interactions dominate the field
Over half of the studies focused on DDIs, reflecting both the availability of data and the seriousness of these interactions. Here, graph-based models (like Graph Neural Networks) and deep learning methods have become increasingly popular, outperforming older machine learning approaches in accuracy. -
Deep learning adds depth, but at a cost
While deep learning excels at capturing complex patterns, it also suffers from problems like data scarcity, overfitting, and limited interpretability. Researchers are beginning to integrate knowledge graphs and explainable AI tools to make models more trustworthy for clinical use. -
Other interaction types are underexplored
Drug–food, drug–supplement, and drug–allergy interactions remain surprisingly underrepresented, even though they are clinically important. Similarly, drug–microbiome interactions—where gut bacteria activate or deactivate drugs—are an emerging but still sparse research area. -
The rise of large language models (LLMs)
Recent studies are beginning to apply LLMs like BioBERT and GPT-style models to process biomedical literature and unstructured clinical notes. By combining these with structured knowledge graphs, researchers are bridging data gaps and creating more robust, interpretable systems. -
Challenges remain
-
Data imbalance: Most datasets capture positive interactions but not well-documented negatives.
-
Noise and heterogeneity: Clinical records, literature, and pharmacological data often conflict.
-
Explainability: Clinicians need transparent models they can trust, not black boxes.
-
Underrepresentation: Nutrient- and allergy-related interactions lack strong datasets.
Why this matters: Laying the foundation for Egypt HealthGPA
This review is more than an academic exercise. It represents the first building block of the Egypt HealthGPA project, which aims to create a national AI-powered drug interaction checker tailored to local and global healthcare needs.
By mapping the landscape, we identified which approaches are most promising, which datasets are most reliable, and where new innovation is needed. This gives us a blueprint for developing a GPA-powered clinical tool that can:
-
Warn doctors and pharmacists about potential dangerous drug interactions in real time.
-
Incorporate nutritional and disease context, not just drug–drug interactions.
-
Support personalized medicine by integrating genomics, microbiome, and lifestyle data.
-
Use explainable AI so that predictions can be trusted and acted upon in clinical practice.
The road ahead: From review to implementation
Our next steps are clear:
-
Building integrated datasets – pulling together drug, nutrient, disease, and microbiome data to reflect the true complexity of interactions.
-
Developing multimodal AI models – combining graph-based reasoning with LLMs for a hybrid system that is both powerful and interpretable.
-
Focusing on underexplored interactions – especially drug–nutrient and drug–allergy cases, which are highly relevant for everyday healthcare.
-
Deploying efficient, scalable models – exploring Small Language Models (SLMs) for real-time, privacy-preserving predictions that can work even in resource-limited healthcare settings.
Conclusion: Toward safer and smarter healthcare
Drug interactions are one of the most preventable causes of harm in medicine—but only if we can anticipate them. AI offers the tools to make this possible, and our review lays the groundwork for moving from fragmented research to integrated, clinical-grade solutions.
Through Egypt HealthGPA, we envision a future where an AI-powered checker sits at the heart of prescribing workflows—alerting clinicians, empowering patients, and making healthcare safer, more efficient, and more personalized.
Our systematic review is the first step in this journey: a map of where AI in drug interaction prediction stands today, and a compass pointing to the future.
Follow the Topic
-
Journal of Cheminformatics

Journal of Cheminformatics is an open-access journal publishing original peer-reviewed research in all aspects of cheminformatics and molecular modelling.
Related Collections
With Collections, you can get published faster and increase your visibility.
Evaluating AI and machine learning models in cheminformatics: benchmarking techniques and case studies
Journal of Cheminformatics is inviting submissions for a collection on "Evaluating AI and machine learning models in cheminformatics: benchmarking techniques and case studies." This collection aims to advance the field by presenting benchmarking methodologies and case studies that highlight the use of AI and machine learning models in cheminformatics. We seek contributions that provide detailed evaluations of computational tools, innovative benchmarking frameworks, and comprehensive case studies demonstrating the effectiveness of these models in predicting chemical properties. Possible topics can include:
• Novel methodologies for benchmarking AI and machine learning models
• Evaluation and comparison of generative models for de novo drug and material design
• Biases in benchmark datasets: unbiased dataset design, biases in used datasets, bias quantification
• Performance metrics for assessing model robustness and reliability
• Calibrated predictions of errors, including in the presence of distribution shifts
• Development and use of curated validation datasets to test external predictivity
• Comparative analysis of molecular fingerprints and descriptors in virtual screening
• Case studies showcasing successful applications of benchmarking techniques in cheminformatics
• Challenges and future directions in the benchmarking of cheminformatics models
We seek original research articles, reviews, and case studies that provide insights into this topic.
All submissions in this collection undergo the journal’s standard peer review process. Similarly, all manuscripts authored by a Guest Editor(s) will be handled by the Editor-in-Chief. As an open access publication, this journal levies an article processing fee (details here). We recognize that many key stakeholders may not have access to such resources and are committed to supporting participation in this issue wherever resources are a barrier. For more information about what support may be available, please visit OA funding and support, or email OAfundingpolicy@springernature.com or the Editor-in-Chief.
Publishing Model: Open Access
Deadline: Apr 16, 2026
Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in