Demethylation of lignin-derived syringol? There's (now) a peroxygenase for that.
Published in Chemistry, Protocols & Methods, and Cell & Molecular Biology

The inspiration
This project started as a conversation between structural biochemist Whelan and chemist Bell in a stairwell; having identified a need for protein-based sensors of smoke taint molecules in grape juice (Wine Australia; ARC DP230103062 F.W. and K.E.S.; and DP200102411 S.G.B. and others), we thought we could explore the specificity of cytochrome P450 enzymes. The timing was right for the start of Harlington’s post-graduate research project with Whelan and Shearwin, and cytochrome P450s with known affinity for smoke taint molecule guaiacol had recently been reported.
Plant processing from forestry and agriculture generates approximately 100 million tons of waste each year, mostly made up of the hard polymers that form the mechanical support in hard and softwoods, termed 'Lignin'. Currently, only about 2% of lignin waste is processed to recover useful materials. Lignin is one of the most abundant polymers on earth and is an untapped source of diverse chemicals that could be funnelled to production of goods like fragrances, flavourings, fuels and therapeutics. Industrial chemists apply high temperatures, high pressure, strong acids and poisonous solvents to break up the polymers and extract the valuable compounds trapped in the waste to ‘valorise’ lignin. But we've asked, what if we could find a greener, simpler alternative? Fungi and bacteria can use lignin as an energy source—how do they do it?
Mining bacteria for plant waste processing enzymes
Bell had identified two genes encoding proteins in a soil-dwelling bacterium, Amycolatopsis thermoflava, one of which (GcoA) was likely to bind guaiacol (Mallinson et al., Nat. Comms, 2018; Fetherolf et al., PNAS, 2020); the other was unknown, but we hypothesised it could bind syringol given the plant-based constituents of lignin. Both enzymes from this bacterium were produced by recombinant expression and purification; Harlington undertook a comprehensive search for potential substrates, accompanied by the smells of mint, smoke and vanilla wafting around the lab. As predicted, GcoA was observed to bind guaiacol; while the second enzyme appeared to bind syringol and derivatives thereof, with selectivity observed between the two enzymes. This was an exciting observation – to our knowledge, no other native syringol binding Cytochrome P450s had been observed to date. Sequence analysis placed both genes in the CYP255 subfamily and with the syringol specificity in hand, we coined the name SyoA.
Other differences in these two enzymes were observed; they were associated with distinct genes predicted to encode proteins for divergent electron transfer to the P450 to support monooxygenase activity, and catalytic turnover of substrates. We set out to investigate this, but it was a complex undertaking, with unproductive uncoupling side reactions observed to dominate with syringol-based substrates. The complexity of reconstituting this electron-transfer/monooxygenase system led us to investigate other options to achieve monooxygenase catalysis.
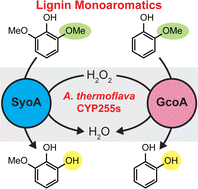
Simplifying catalysis with the peroxide shunt
We examined the sequences of the GcoA and SyoA CYP255 enzymes and observed an unusual pair of amino acid residues at an important location in the enzyme, typically associated with dioxygen activation. We proposed these residues (an acidic glutamate and neutral glutamine) may enable the enzymes to use hydrogen peroxide, rather than dioxygen, to generate their reactive intermediate for catalytic function (Fig. 1). We demonstrated that this was indeed the case for both enzymes, and that both were relatively stable in the presence of hydrogen peroxide (Harlington et al., 2022). We then set out to discover the active site structures that enable these enzymes to function as peroxygenases rather than monooxygenases.
We determined the structure of SyoA to atomic resolution in the open unbound state. Considerable effort in co-crystallisation with substrate was invested to determine the atomic-resolution syringol-bound state of the closed conformation of SyoA (Fig. 2). This illustrated that the active site had clear space to accommodate a para-substituted substrate. Having a methyl-substituent at the para position of syringol defined the highest affinity binder.

Fig. 2 Slab render of SyoA bound to substrate syringol (yellow/red) and cofactor heme (white).
Structures of SyoA with substrate bound enabled high-resolution definition of the open to closed transition of the enzyme; and the relative atomic positions of substrates, heme cofactor and catalytic residues. In comparison to other substrate-bound P450s, key differences in the position of residues important for generating the iron-oxygen intermediates were observed. The functional importance of these residues was interrogated by site-directed mutagenesis by Harlington and post-graduate student Das, with impacts on substrate-binding and catalytic turnover assessed.

Specific findings with broad applicability
- This work is important for understanding bacterial degradation of lignin-derived aromatics in their biological context. It is also pivotal in understanding and simplifying the application of heme enzymes as biocatalysts.
- We have used the information obtained here to engineer peroxygenase activity into other P450 enzymes, simplifying their application and enabling the generation of active and highly selective and thermostable biocatalysts for C-H bond functionalization reactions in the chemical synthesis space (Das et al., 2025; Gee et al, 2023).
- In the future, these biocatalysts can be used to design biocatalytic functions into other artificial and natural heme proteins/enzymes.
- The conversation in the stairwell still resonates in the potential of these enzymes as sensors of lignin-derived monoaromatics.
- Value-added prospect of utility for degradation and valorization of other lignin derived aromatics, either directly for syringol and guaiacol, or other engineered P450 systems exploiting this work on peroxide-driven P450 activity.
Das, T., Hayball, E.F., Harlington, A.C., and Bell, S.G. A Thermostable Heme Protein Fold Adapted for Stereoselective C−H Bond Hydroxylation Using Peroxygenase Activity. ChemBioChem 26, e202400737 (2025).
Fetherolf, M.M., Levy-Booth, D.J., Navas, L.E. and Eltis, L.D. Characterization of alkylguaiacol-degrading cytochromes P450 for the biocatalytic valorization of lignin. Proc. Natl. Acad. Sci. USA 117, 25771–25778 (2020).
Gee, A.R., Stone, I.S.J., Stockdale T.P., Pukala, T.L., De Voss J.J., S.G. Bell Efficient biocatalytic C–H bond oxidation: an engineered heme-thiolate peroxygenase from a thermostable cytochrome P450. Chemical Communications 59(90), 13486-13489 (2023).
Harlington, A.C., Shearwin, K.E., Bell, S.G. and Whelan F. Efficient O-demethylation of lignin monoaromatics using the peroxygenase activity of cytochrome P450 enzymes. Chemical Communications 58, 13321-13324 (2022).
Mallinson, S.J.B., et al. A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion. Nat. Commun. 9, 2487 (2018).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026




Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in