Developing a therapeutic RNA base editing platform
Published in Bioengineering & Biotechnology
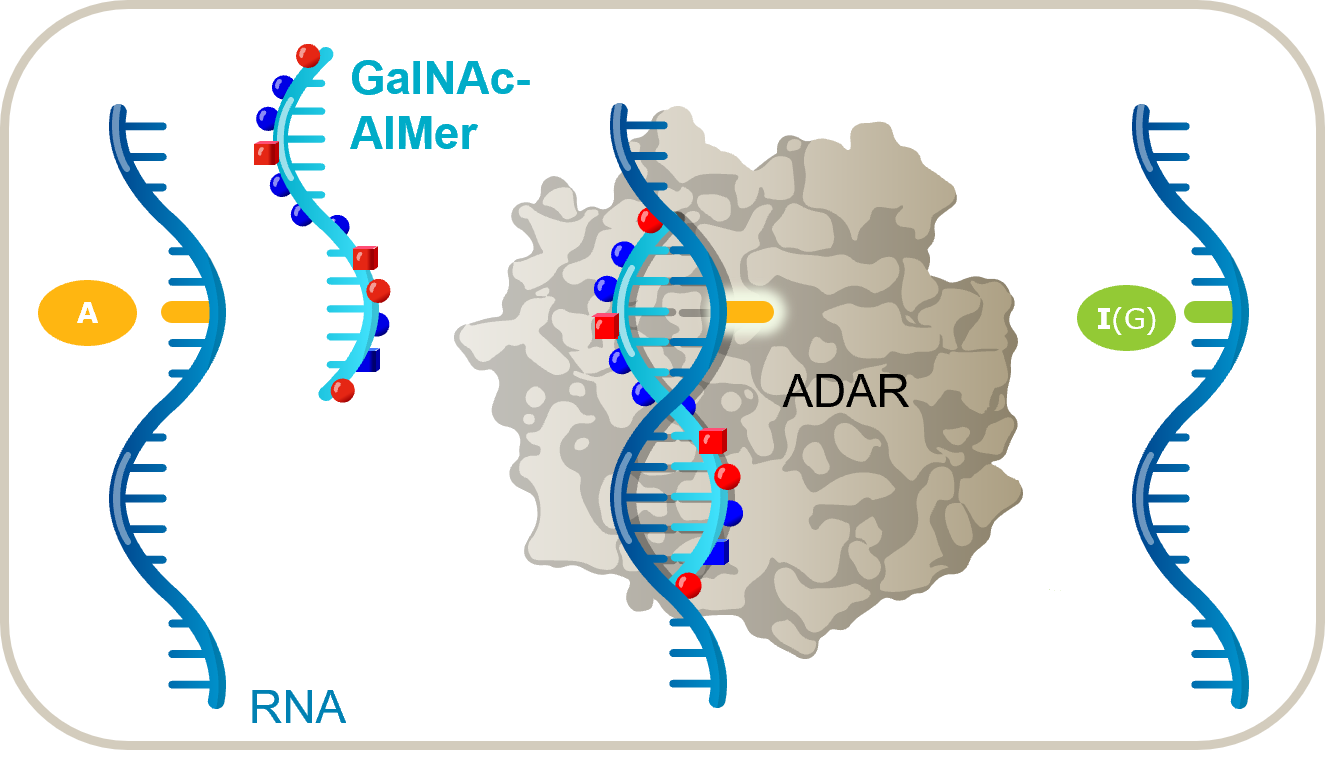
Our scientific team, led by Dr. Chikdu Shivalila and Dr. Prashant Monian, set out to create a therapeutic platform that supported RNA base editing by recruiting ADAR (adenosine deaminases acting on RNA) enzymes. Our goals were straightforward but challenging:
- to build a platform based on chemically modified stereopure oligonucleotides that is comparable in simplicity to other therapeutic antisense technologies such as exon skippers or gapmers that promote RNase H-mediated silencing
- to keep the oligonucleotides short so they do not require ancillary technologies to facilitate delivery (e.g., viral vector, lipid nanoparticles)
- to recruit endogenous enzymes with the intention of avoiding the off-target editing and delivery challenges associated with exogenous ADAR
- to potentially overcome barriers to translating RNA editing efficiency from in vitro to in vivo, which have thwarted prior technologies in this field
|
Dr. Chikdu Shivalila |
Dr. Prashant Monian |
|
Dr. Chikdu Shivalila is a Principal Scientist at Wave. He earned his PhD in Biological Sciences in the laboratory of Dr. Rudolf Jaenisch at MIT/Whitehead Institute of Biomedical Sciences, where he focused on genome and epigenetic engineering. When Chikdu and the team commenced work to develop an RNA base editing modality at Wave, they began by throwing out the rule book, discarding the notion that double-stranded guide RNAs were necessary. The team believed from the very beginning that editing could be achieved with short, chemically modified, single-stranded oligonucleotides. |
Dr. Prashant Monian is a Senior Scientist at Wave. He earned his PhD in Biomedical Sciences in the laboratory of Dr. Xuejun Jiang at the Gerstner Sloan-Kettering Graduate School of Biomedical Sciences at Memorial Sloan-Kettering Cancer Center, where he worked to elucidate the molecular basis for programmed cell death through apoptosis and ferroptosis. Seeking to get closer to research in the biotech industry, Prashant joined Wave as a post-doctoral fellow and soon began work on establishing an RNA base editing platform for the company. |
To achieve the established goals for delivering an RNA base editing modality with therapeutic potential, our scientists leveraged knowledge coming from a decade of research at Wave exploring how chemical modifications, including backbone chemistry and stereochemistry, interact with base sequence to impact the pharmacology and biological mechanisms of oligonucleotides1-5.
Wave was founded on the notion that control of backbone stereochemistry represented an opportunity to improve the therapeutic potential for chemically modified oligonucleotides. This notion, which came from our founders Professors Gregory Verdine and Takeshi Wada, inspired our investment in and exploration of the complex relationships between backbone chemistry and stereochemistry and other features of an oligonucleotide (e.g., base sequence, sugar modifications). These investigations, which spanned multiple oligonucleotide modalities, formed the foundation for PRISMTM, Wave’s discovery and drug development platform.
In their application of PRISM to RNA base editing, our team began with a simple hypothesis: natural ADAR substrates resemble a duplex between an RNA-based oligonucleotide and an endogenous RNA (Figure 1). If correct, this meant that it should be possible to design oligonucleotides that interact with transcripts through complementary base pairing to form a double-stranded RNA substrate that, in turn, recruits and directs the editing activity of endogenous ADAR enzymes.
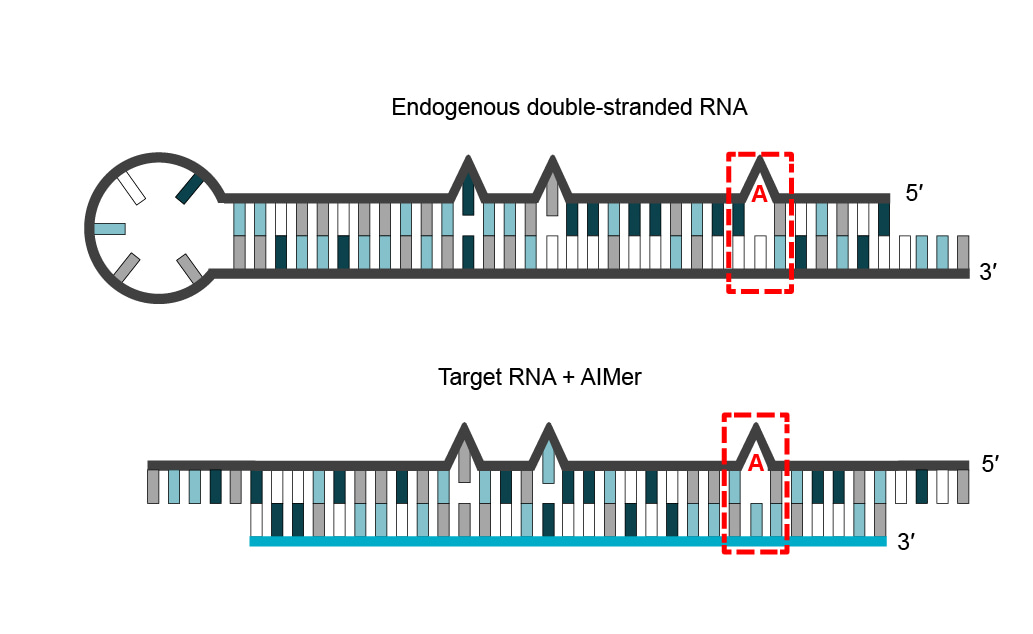
Figure 1 | Endogenous substrates for ADAR editing (top) resemble a duplex (bottom) between an RNA-based oligonucleotide (AIMer, blue backbone) and an RNA substrate (black backbone).
Although the team had insight into how sequence, chemistry, and stereochemistry impacted the pharmacology of oligonucleotides in general from PRISM, we lacked specific insight into how these features, including our novel stereopure phosphoryl guanidine (PN) backbone chemistry, would interact with and impact ADAR enzymes. To understand these relationships, we needed to establish an in-house screening system for quick and efficient assessment in cells. The team identified a highly sensitive reporter system that allowed for detection of low levels of activity in cells. It incorporated a gain-of-function component that required A-to-I editing of a luciferase gene to provide insight into activity (Figure 2). Importantly, this system was also suited for measuring editing at scale and over time, allowing us to make the most of our unique ability to generate and evaluate the activity of hundreds of stereopure oligonucleotides in a very short time thanks to process development achievements realized through PRISM.
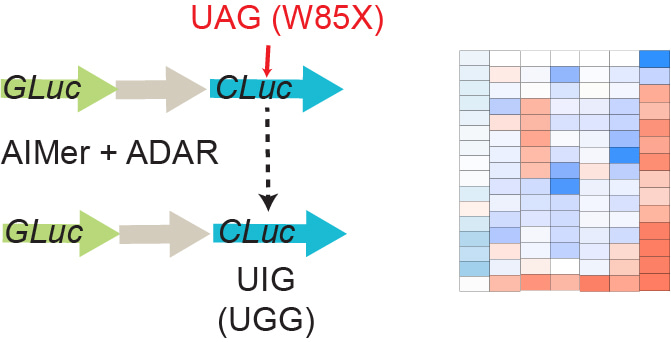
Figure 2 | To optimize key design features, we evaluated hundreds of stereopure chemically modified oligonucleotides in a cellular luciferase reporter system.
Accordingly, our team generated and began testing oligonucleotides designed to methodically interrogate the relationship between specific chemical modifications and RNA base editing. Within months, they observed oligonucleotide-dependent RNA base editing. This observation validated the overall approach and kick-started Wave’s extensive efforts to understand how ADAR-dependent RNA base editing activity could be improved through Wave’s PRISM platform.
In our initial studies, our team also evaluated multiple types of ADAR enzymes, including those derived from different model organisms, which proved critical. This strategy enabled us to optimize key design features for our editing oligonucleotides, which we called AIMers to highlight their A-to-I editing activity and the fact that they are short (mers).
We next applied these AIMer design features to multiple endogenous transcripts harboring the base targeted for editing in different locations along the transcript and in different sequence contexts. We also confirmed AIMers were compatible with GalNAc conjugation. These applications helped ensure our AIMer technology would be versatile, rendering it a viable option for various therapeutic RNA editing opportunities. We also confirmed that we could safely co-opt endogenous ADAR machinery to support AIMer directed RNA base editing without compromising the natural homeostatic editing activity of ADAR enzymes.
Finally, we advanced AIMers into the types of experiments that have emerged as roadblocks for other technologies: translation to animal models and specificity of activity. For the former challenge, we realized an unexpected benefit from our expansive screen to understand the relationship between chemical modification and the resulting ADAR editing activity. During the course of these experiments, we noted that the same ADAR enzyme from different species (mouse Adar1 and human ADAR1) showed different activity with the same modifications for various targets. This observation helped us appreciate that there are likely species-specific differences in how ADAR enzymes recognize substrates or catalyze base editing that have not been fully appreciated. We have also learned from our other programs that the application of PN chemistry has a profound and beneficial impact on the activity of our oligonucleotides in preclinical in vivo models 4, 5. Because of these observations, we opted to test our PN-containing AIMers, which were optimized for human ADAR enzyme activity in vitro, in non-human primates rather than in mice. We believe this decision was a key factor in our ability to rapidly and efficiently translate our AIMer technology in vivo, with multiple GalNAc-conjugated AIMers yielding ~50% editing of ACTB in the NHP liver.
For specificity, we felt confident that by leveraging endogenous enzymes that remain under physiological control, we would be facing fewer challenges than technologies that overexpress exogenous and sometimes hyperactivated ADAR6-12. Indeed, this hypothesis proved true, as none of the AIMers we tested during the course of this work elicited the type of off-targeting or bystander editing that has become an Achille’s heel for other editing technologies.
Since the completion of experiments reported in this publication, we have continued to explore the relationship between the chemistry and/or structure of an AIMer-RNA substrate to improve our AIMer designs. We have launched a therapeutic discovery program in Alpha-1 antitrypsin deficiency (AATD). We are developing AIMers to correct an E342K missense mutation in the SERPINA1 mRNA, which encodes a mutant form of AAT, called Z-AAT, that aggregates in hepatocytes, decreasing the secretion of functional AAT in serum and resulting in a gain-of-toxicity in liver and loss-of-AAT-function in the lung13. We report preliminary work from this discovery-stage program in the paper, showing that AIMers can direct RNA base editing in SERPINA1 and that this editing increases secretion of functional AAT from hepatocytes in vitro.
This paper reports Wave’s foundational efforts to build a therapeutic RNA base editing platform. It illustrates the vision of our founders, the innovative spirit of our scientists, and the value added from our long-term investments in PRISM and the processes that support stereopure oligonucleotide synthesis.
Read the paper here: https://www.nature.com/articles/s41587-022-01225-1
More information about Wave Life Sciences: https://wavelifesciences.com/
References
- Iwamoto, N. et al. Control of phosphorothioate stereochemistry substantially increases the efficacy of antisense oligonucleotides. Nat Biotechnol 35, 845-851 (2017).
- Liu, Y. et al. Variant-selective stereopure oligonucleotides protect against pathologies associated with C9orf72-repeat expansion in preclinical models. Nat Commun 12, 847 (2021).
- Byrne, M. et al. Stereochemistry Enhances Potency, Efficacy, and Durability of Malat1 Antisense Oligonucleotides In Vitro and In Vivo in Multiple Species. Transl Vis Sci Technol 10, 23 (2021).
- Kandasamy, P. et al. Control of backbone chemistry and chirality boost oligonucleotide splice switching activity. Nucleic acids research (2022).
- Kandasamy, P. et al. Impact of guanidine-containing backbone linkages on stereopure antisense oligonucleotides in the CNS. Nucleic acids research (2022).
- Montiel-Gonzalez, M.F., Vallecillo-Viejo, I., Yudowski, G.A. & Rosenthal, J.J. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci U S A 110, 18285-18290 (2013).
- Fukuda, M. et al. Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. Sci Rep 7, 41478 (2017).
- Vallecillo-Viejo, I.C., Liscovitch-Brauer, N., Montiel-Gonzalez, M.F., Eisenberg, E. & Rosenthal, J.J.C. Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol 15, 104-114 (2018).
- Sinnamon, J.R. et al. Site-directed RNA repair of endogenous Mecp2 RNA in neurons. Proc Natl Acad Sci U S A 114, E9395-e9402 (2017).
- Cox, D.B.T. et al. RNA editing with CRISPR-Cas13. Science 358, 1019-1027 (2017).
- Montiel-González, M.F., Vallecillo-Viejo, I.C. & Rosenthal, J.J. An efficient system for selectively altering genetic information within mRNAs. Nucleic acids research 44, e157 (2016).
- Katrekar, D. et al. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat Methods 16, 239-242 (2019).
- Strnad, P., McElvaney, N.G. & Lomas, D.A. Alpha(1)-Antitrypsin Deficiency. N Engl J Med 382, 1443-1455 (2020).
Follow the Topic
-
Nature Biotechnology

A monthly journal covering the science and business of biotechnology, with new concepts in technology/methodology of relevance to the biological, biomedical, agricultural and environmental sciences.





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in