Differential inheritance of gene content among maize, rice, wheat, and bamboos following ancestral duplications and possible contribution to morphological and physiological differences
Published in Ecology & Evolution, Genetics & Genomics, and Plant Science
Major crops such as maize, wheat, rice, as well as bamboos, are all members of the grass family (Poaceae) , yet they exhibit distinct morphologies and various physiological properties, such as adaptation to different environments. Poaceae are the fifth-largest family of flowering plants, ~12,000 species classified into 12 subfamilies. The largest subfamily is Pooideae; in addition to wheat, other Pooideae members include barley, oat, ryegrass, orchardgrass, brome, and annual meadow-grass. Maize belongs to Panicoideae, the second largest; moreover, sorghum, foxtail millet, sugarcane, fountaingrass, and switchgrass and also in this subfamily. Domesticated and wild rice species and their close relatively form the subfamily Oryzoideae, which bamboos belong to Bambusoideae, which are the third largest. Another major subfamily, Chloridoideae, include the crop teff and other grasses: sea oats, saltgrass, and finger grass.
Although grasses share a number of morphological characteristics, they also display wide morphological variations, such as plant heights, leaf shapes, and inflorescence architectures. Specifically, most grasses are herbaceous, but bamboos are most woody. Grasses also vary dramatically in their adaptation to different environmental conditions, including temperature and water availability. For example, rice and other Oryzoideae members grow in aquatic environments, whereas most other grasses are found in dry ecosystems. In addition, wheat and other Pooideae members are adapted to high latitudes and altitudes with low temperatures, but grasses in Panicoideae and Cloridoideae are distributed in areas with hot and dry environments, often with the adaptive C4 photosynthesis.
Polyploid plants are widely spread, such as the tetraploid cotton, tetraploid potato, triploid banana, hexaploid wheat, and octoploid strawberry, as well as many in nature. These polyploid plants underwent relatively recent whole genome duplications (or increases) and exhibit often increased yields (such as larger size) and enhanced adaptability to diverse stresses (such as disease resistance) compared with their diploid relatives. Genomic and phylogenomic studies have supported the idea that angiosperms share a whole genome duplication (WGD), with additional WGDs detected at the origins of large groups of angiosperms. During evolution, numerous duplicate genes from WGDs have been retained, whereas most are lost. The retained duplicates can further increase in copy number after further WGDs in some lineages, providing important genetic materials for functional innovation and species evolution.
Specifically, grass genomes have been found to shared a common WGD event, named rho, which was linked to the increase of diversification rate in early Poaceae. However, genome-wide analyses of the number and potential functional differentiation of rho-derived gene duplicates have not been conducted to detect retention and loss patterns using multiple representatives in different subfamilies, largely because genome sequences for several Poaceae subfamilies were not available until very recently. With help and contributions from many colleagues and botanical gardens, our lab assembled >300 large-scale grass sequence datasets, including transcriptomes and genome-skimming datasets. These were combined with additional public genomic/transcriptomic datasets to represent all subfamilies to construct highly resolved nuclear species-trees of Poaceae (Huang et al., 2022 Molecular Plant) and Pooideae (Zhang et al., 2022 Molecular Biology and Evolution).
Using the nuclear phylogeny as a reference and 363 genome-scale grass datasets, we conducted phylogenomic and phylotranscriptomic investigation of WGDs in multiple grass subfamilies, identifying evidence for previously unknown WGDs and for gene conversion of rho duplicates and implicating duplicates in environmental adaptations or morphogenesis. Specifically, we found that some of rho-derived duplicates correspond to the core Poaceae other than the grass ancestor. These and other duplicates apparently experienced sequence equalization likely due to gene conversion. We also found that 16 WGDs are placed in the phylogenetic clades with high species richness, suggesting possible contribution of WGDs to the evolution of these highly diverse clades. In addition, we found the retention of the rho-derived duplicates related to agricultural trait or adaptations in one subfamily, but lost in other subfamilies, including the rice GROWTH-REGULATING FACTOR 4 gene and the maize RAVL1 gene. Among the rho-duplicates retained in the most recent common ancestor of grass, most have experienced reciprocal losses and/or duplications among subfamilies, such as the genes GRF4 (Duan et al., 2015 Nature Plants), KRN2 (Chen et al., 2022 Science), and DREB1C (Wei et al., 2022 Science).
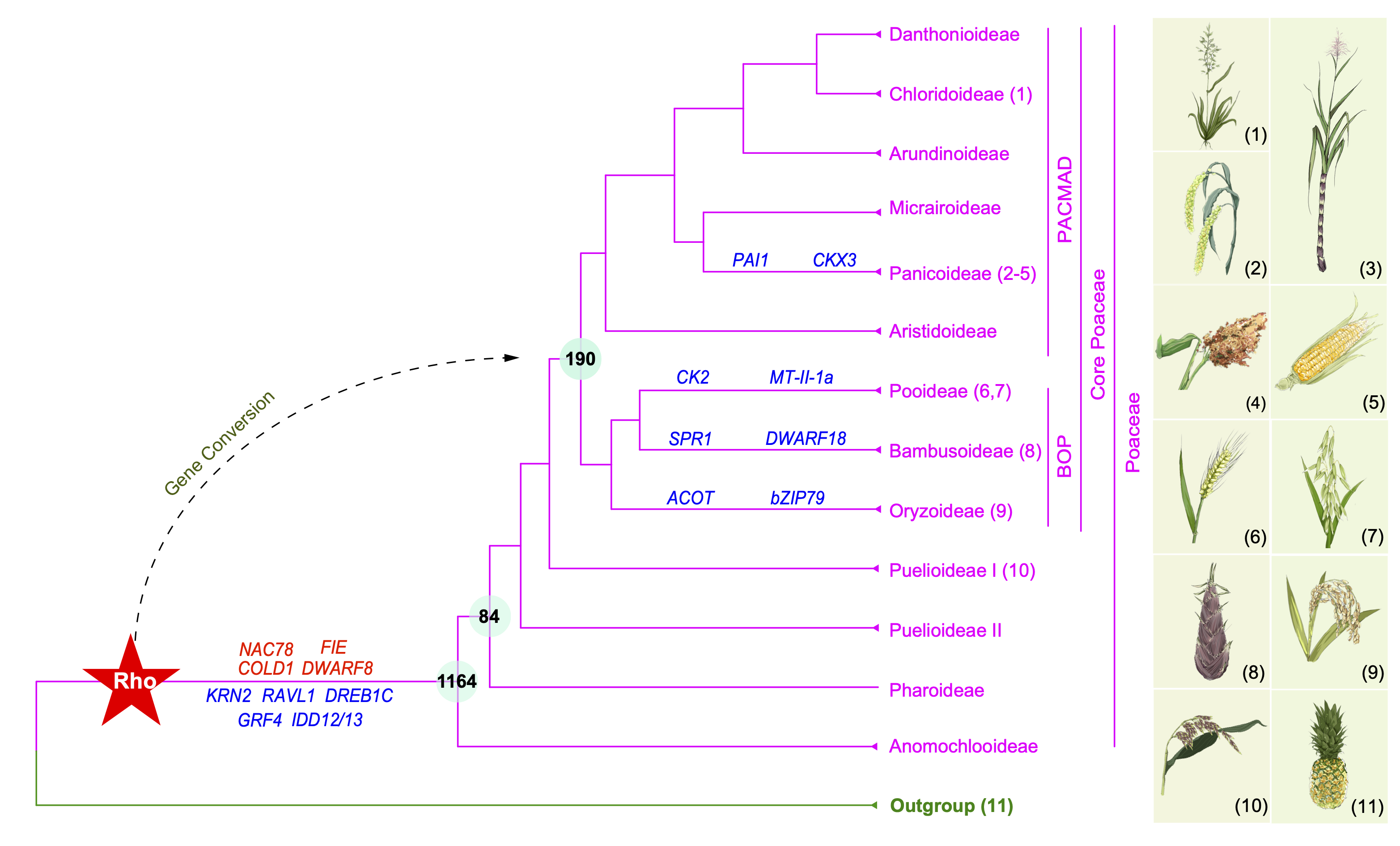
The figure was adapted from our published Figure 2 and modified according to genes shown in the published Tables 1 and 2. 1-teff, Eragrostis tef; 2-foxtail millet, Setaria italica; 3- sugarcane, Saccharum spontaneum; 4-sorghum, Sorghum bicolor; 5-maize, Zea mays; 6-wheat, Triticum aestivum; 7-oat, Avena sativa; 8-bamboo, Phyllostachys edulis; 9-rice, Oryza sativa; 10- Puelia ciliata; and 11-pineapple, Ananas comosus.
It is thought that WGDs often resulted from ancient hybridizations. However, the parental lineages of many well-characterized WGDs are unknown, including rho. We found support for a WGD shared by woody bamboos and for herbaceous bamboos being related to a parental lineage of woody bamboo. We further identified gene phylogenetic support for a hybridization in the Oryza genus, with domesticated and wild rice, for the origin of the tetraploid Oryza coarctata, with one parent related to the previously defined F genome in Oryza and the other previously unknown genome similar to the ancestor of the A, B, C, and E genomes.
The view from the first author, Dr. Taikui Zhang:
I am a postdoctoral scientist in Dr. Ma’s lab and performed the bioinformatic and molecular evolutionary analyses of grass datasets in this study. The great grass diversity has stimulated my interests in exploring the grass genome evolutionary patterns and their biological implications. Some of the strategies that I have learned from Dr. Ma is to divide a complex task into several component tasks and to complete the more feasible task before others. Also, when we detected evidence for WGD events from different approaches, we prioritized the further exploration of phylogenomic results because it could provide additional information of the placement of many gene duplications at different nodes of the grass species-tree and revealed new insights into gene conversion. WGDs are associated with many large families and other large groups of angiosperms; thus our findings of the processes for lineage-specific and WGD-related gene evolution can have general implications throughout the angiosperm history.
The view from corresponding author, Dr. Ji Qi:
The Poaceae family, commonly known as grasses, harbors a vast spectrum of species with notable ecological and economic importance. From the important crops to rapid growing bamboos, Poaceae species have adapted to diverse habitats and environmental conditions, exhibiting a remarkable array of morphologies and physiological traits. Within the vast landscape of Poaceae evolution, whole-genome duplications stand out as important events, shaping the genetic architecture and evolutionary trajectories of grass species. These WGD events, which generate copies of all genes including those involved in the same networks or pathways, offer greater potential for the neo/subfunctionalization of duplicates through the co-evolution of functionally related genes. Through comprehensive phylogenomic and transcriptomic analyses, our investigation reveals that the Poaceae family have undergone multiple polyploidization events before and after sub-lineage divergence. In addition, the progeny species exhibit large scale gene conversion events, potentially contributing to the increasing of genome complexity and species diversity. By elucidating the patterns of preferential retention and loss of duplicated genes among different sub-families, our findings provide clues for understanding the mechanisms driving lineage-specific adaptations and speciation events within grasses.
The view from corresponding author, Dr. Hong Ma:
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in