Differential response of antibiotic therapy to the infant and adult gut microbiome
Published in Microbiology

Our team has been studying infant gut microbiome in the past few years1-3, with a special focus recently on how antibiotic therapy affects health via microbiome. Antibiotics are a two-edged sword: while antibiotics are crucial for fighting pathogenic bacteria, they can also lead to resistance and disrupt the delicate balance of gut flora. We are interested in understanding the negative consequences of antibiotics, especially the disparities in how these effects express themselves in different age groups. We believe that differences in the composition of gut microbial communities at different ages may lead them to show different stability and resilience in the face of external environmental disturbances, such as antibiotic treatment. Since 2021, we have launched an in-depth investigation and study of two different age groups, adults and infants, to understand how the gut microorganisms respond to antibiotic stress and how this phenomenon varies across age groups.
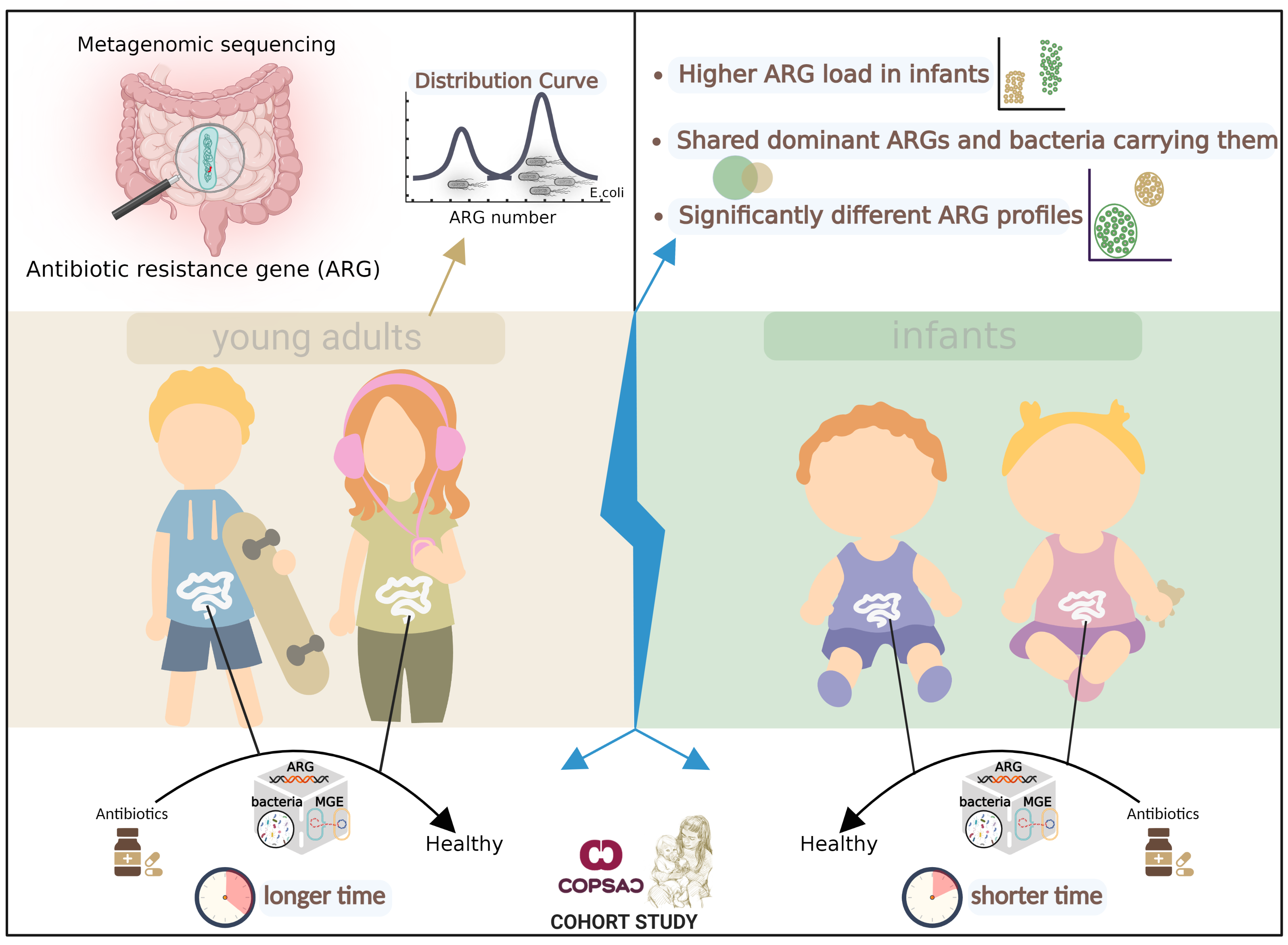
Fig. 1 Overview of Key Findings. We sequenced fecal metagenomes from COPSAC cohort of 217 adults aged 18 years and 662 infants aged one year. The distribution of resistance gene richness among samples was bimodal, with one peak of low richness and another of high richness, driven by E. coli composition. The profiles of resistance genes were significantly different between adults and infants and antibiotic resistance genes were more abundant in the infant guts than in adult guts. Strikingly, Infants and adults shared dominant ARGs and bacterial species carrying them in the gut microbiome. Compared to infants, antibiotic treatment in adults had a longer-lasting effect on microbial composition, resistance gene and Mobile genetic element profiles, and plasmid abundance. The total abundance of core ARGs—resistance genes that are highly abundant and prevalent overall—also increased in the guts of both adults and infants after antibiotic treatment.
Escherichia coli and antibiotic resistance genes:
A bimodal distribution of antibiotic resistance genes was observed in both adult and infant cohorts (Fig. 1). In these cohorts, one group exhibited high levels of antibiotic resistance genes in their guts, while the other group showed low levels. This distribution was mainly influenced by E. coli abundance and was not related to age. The extremely wide range of antibiotic resistance in Enterobacteriaceae bacteria is largely attributable to gene flow under sustained selective pressure resulting from increased antibiotic use in recent decades. The similar profiles of resistance genes in Enterobacteriaceae strains in both adult and infant guts further suggest frequent gene influx in these bacterial genomes.
Age differences and resistance gene similarity:
The loads of antibiotic resistance genes in the guts of infants were significantly higher compared to those in adults (Fig. 1). This can be attributed mainly to the higher levels of E. coli and mobile vector plasmids found in the infant gut. In this study, we found that the dominant antibiotic resistance genes and their carrier strains were very similar in the guts of infants and adults, despite age differences (Fig. 1). The main differences in resistance genes between them came from genes with lower abundance. This raises the question: do these genes and bacteria establish a sustained selective advantage or a shared community pool early in life? This question triggers deeper thinking about antibiotic use in infancy, external environmental selection, and host immune system regulation.
Cross-age effects of antibiotics on gut microbes:
We explored the perturbation of the gut microbiome by antibiotics in the treatment of routine infections (Fig. 1). Strikingly, the gut microbiome of infants responded more quickly to antibiotics (~1 month), whereas adults took longer to recover (~1 year). Four assessments – microbial/mobile genetic element compositions, antibiotic resistance gene profiles, and plasmid abundance - consistently showed these trends in the gut microbiome. This could mean that although the infant gut microbiome is more fragile, it also implies greater resilience and recovery. The next interesting finding was that core resistance genes (high prevalence and abundance genes) were significantly affected by antibiotic treatment in both populations. This means that they may be the main weapon of bacteria against antibiotics and therefore may be widely spread among bacteria, as seems to be evidenced by their high prevalence and abundance patterns.
Long-term effects of antibiotic therapy:
Although the infant's gut microbiome may return to baseline levels within approximately 30 days, it remains to be explored whether there are potential long-term effects on specific resistance genes, immune system maturation, or metabolic changes. This gives us direction for future in-depth studies.
The role of different antibiotics:
We found that different types of antibiotics have different effects on antibiotic resistance genes and mobile genetic elements in the gut microbiome. In adults, tetracycline and β-lactam plus macrolide antibiotics showed the greatest effect on antibiotic resistance genes and plasmids. In infants, penicillin and macrolide antibiotics demonstrated a strong association with high antibiotic resistance gene abundance. This reflects the varying selectivity and impact of different antibiotics on the microbiome.
Limitations of the study and plans for further research:
Only two age groups were included in our study, which means we have not yet fully understood how antibiotics affect gut microbes across age groups. However, this points us in the direction of future research: We intend to collect additional data to further explore and understand the effects of antibiotics on gut microbes across a broader range of age groups.
References
- Li, X., Stokholm, J., Brejnrod, A., Alberg Vestergaard, G., Russel, J., Trivedi, U., Thorsen, J., Gupta, S., Hjort Hjelmsø, M., A Shah, S., Arendt Rasmussen, M., Bisgaard, H., & Soerensen, S. J. (2021). The infant gut resistome associates with E. coli, environmental exposures, gut microbiome maturity, and asthma-associated bacterial composition. Cell Host and Microbe, 1–13. https://doi.org/10.1016/j.chom.2021.03.017
- Stokholm, J., Blaser, M. J., Thorsen, J., Rasmussen, M. A., Waage, J., Vinding, R. K., Schoos, A. M. M., Kunøe, A., Fink, N. R., Chawes, B. L., Bønnelykke, K., Brejnrod, A. D., Mortensen, M. S., Al-Soud, W. A., Sørensen, S. J., & Bisgaard, H. (2018). Maturation of the gut microbiome and risk of asthma in childhood. Nature Communications. https://doi.org/10.1038/s41467-017-02573-2
- Stokholm, J., Thorsen, J., Blaser, M. J., Rasmussen, M. A., Hjelmsø, M., Shah, S., Christensen, E. D., Chawes, B. L., Bønnelykke, K., Brix, S., Mortensen, M. S., Brejnrod, A., Vestergaard, G., Trivedi, U., Sørensen, S. J., & Bisgaard, H. (2020). Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Science Translational Medicine. https://doi.org/10.1126/scitranslmed.aax9929
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in