Diversity and Durability in Plant Disease Resistance
Published in Plant Science and Agricultural & Food Science

|
The attractive village of Chidham lies on the coast of West Sussex, England, close to the historic cathedral city of Chichester. In former times, despite its maritime location, the people of Chidham were mostly farmers rather than sailors, as the parish had a good reputation for its rich arable land. It was there that in 1790, a sharp-eyed Mr Woods made an intriguing discovery: a single wheat plant in a hedgerow which he thought could be worth multiplying and marketing. Sales were successful and over the next half-century, Chidham White became widely grown throughout Great Britain. This wheat variety was well-known enough to be mentioned by Darwin in The Variation of Animals and Plants under Domestication in 1868 and in several agricultural publications [1]. |

|
When Louis de Vilmorin founded the world’s first wheat breeding programme around 1860, he naturally used the popular Chiddam à Epi Blanc, as Chidham White was known in France, as one of his founder stocks. What neither Vilmorin nor Darwin nor the observant Mr Woods could have known was that among the many genes that Chidham White would contribute to modern wheat varieties was Stb15, a gene controlling resistance to Septoria tritici blotch, which would not even be recognised as a disease of wheat for another half-century. Its descendants bred in France and Belgium form the genetic backbone of modern European wheat, culminating in 1946 in Cappelle Desprez, one of the most popular wheat varieties ever produced. Chidham White is in the parentage of all wheat grown in Britain, 60% of which has Stb15 [2].
Septoria has become the most severe disease of wheat in humid, temperate regions worldwide. When we decided to study molecular evolution of Septoria resistance, my collaborator Brande Wulff, our student (later postdoc) Amber Hafeez and I chose Stb15 as our target gene because it has an especially clear response to the fungal pathogen, Zymoseptoria tritici [3]. The core of our work to clone and characterise Stb15 was the Watkins Collection, a remarkable resource created nearly a century ago by A.E. Watkins of Cambridge University, who assembled a world-wide collection of over 1000 local wheat landrace varieties [4]. The Watkins Collection is a snapshot of the diversity of wheat as it had evolved over 10,000 years of agriculture, just before most landraces were displaced by new Green Revolution varieties.
 |
There is huge diversity in Septoria resistance among the hundreds of landrace wheat varieties in the Watkins Collection [5]. The black spots on the leaves are pycnidia, the asexual fruiting bodies of the Septoria fungus, Zymoseptoria tritici. |
In a genome-wide association study (GWAS), the sequence diversity over most of the genome, spread over many diverse landraces, was background noise against which the clear signal of allelic variation in Stb15 resistance stood out. This powerfully illustrates how GWAS on a large, diverse set of landraces has created opportunities to clone genes from the huge genome of wheat with astonishing speed and precision. The GWAS materials, combined with multi-disciplinary skills in pathology, genetics, genomics, bioinformatics, transformation and protein modelling, enabled rapid identification of just a few candidate genes followed by validation tests to confirm the identity of Stb15, with an intercontinental network of labs playing parts brilliantly orchestrated by Amber Hafeez [5].
| The diversity of the three Stb genes cloned to date is intriguing. Like Stb6 and Stb16q, Stb15 encodes a receptor-like kinase (RLK) but the three gene products belong to different families. The Stb15 protein is a G-type lectin RLK with an intracellular receptor-like domain and several extracellular domains. How variation in Stb proteins relates to the mechanism of host defence against Septoria is not yet known. Like some other plant diseases such as rusts and mildews, Septoria resistance is controlled by gene-for-gene interactions between the host and pathogen. Genes such as Stb15, as well as Stb6 and Stb16q, greatly enhance a wheat variety’s resistance if the invading Z. tritici fungus has a specific avirulence (Avr) phenotype. For Stb15, this is AvrStb15, although the genetic control of this Avr has not been characterised yet. |
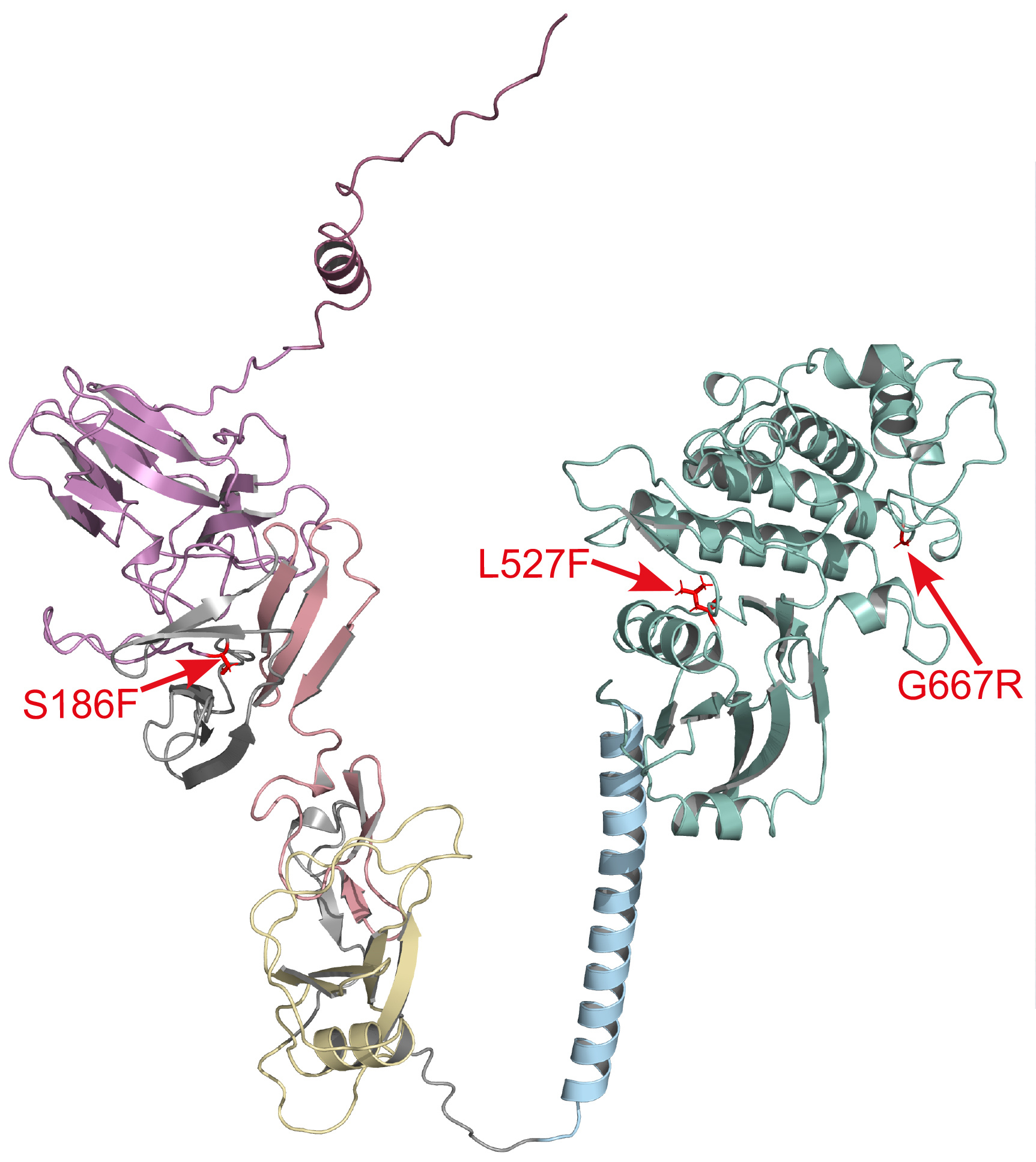
|
Even though scientific understanding of host defence against Septoria is currently limited, breeders have made good progress in producing varieties with improved resistance. For the most part, they have not relied on genes such as Stb15 which control recognition of avirulent Z. tritici. Although recognition-based resistance can be strong, its downside is that it is not durable, because the fungus can evade it by loss or mutation of the Avr protein. That is the case for Stb15 in Europe, where almost the entire Z. tritici population is virulent to it, although Stb15 may have some temporary benefit elsewhere. It seems that, instead, wheat breeders have built up polygenic, quantitative resistance to Septoria by combining many genes with small individual effects through phenotypic selection in breeding nurseries and more recently by genomic selection [6]. This type of resistance is durable because it is effective against all pathogen genotypes. That’s useful because it protects plant breeders’ investment in improving many other traits and means that farmers are not suddenly faced with a substantial loss of grain production caused by a severe outbreak of disease. Scattered evidence from diverse plant diseases suggests that durable resistance largely involves variation in many downstream defences rather than recognition of single Avr proteins [7].
A potential benefit of characterising major Stb genes like Stb15 is an indirect one, because it may help breeders invest in the most advantageous germplasm. When we understand the mechanism of gene-for-gene recognition in Septoria, we may be able to decide if a new source of resistance is unlikely to involve fungal recognition and is thus likely to be durable. There is evidence that Stb-Avr recognition inhibits Z. tritici from penetrating the leaf through the stomata [8]. Does this hold for Stb genes in general, including Stb15? And does durable resistance involve mechanisms other than obstruction of stomatal penetration?
As so often in science, the most useful practical outcome of our research might be something we were not originally looking for. Among the hundreds of Watkins lines we tested, we found tens with high levels of quantitative, background resistance to Septoria. This was not related to interactions with specific fungal isolates and therefore might be durable. Like Chidham White over 200 years ago, Watkins landraces from diverse environments across Eurasia and North Africa may be a valuable resource for wheat breeding. GWAS technology has been immensely powerful in cloning single genes with large effects; can it rise to the challenge of enabling breeders to transfer hundreds of genes with small individual effects on Septoria resistance from wheat lines grown in local subsistence farming to modern varieties with excellent yield and quality in diverse environments worldwide?
 |
Breeders' selections of top wheat varieties are assessed by the Agriculture and Horticulture Development Board in Recommended List trials at around 30 UK sites annually. Over 30 traits are scored, of which the four most important are yield, grain quality, standing power (the ability to withstand bad weather) and Septoria resistance.© AHDB / Shooting Stone Media |
References
[1] BBA Wheat Portal. https://www.wheat-gateway.org.uk/hub.php?ID=38
[2] Arraiano & Brown 2017, Molecular Plant Pathology 18, 276-292. https://doi.org/10.1111/mpp.12482
[3] Arraiano et al. 2007, Plant Pathology 56, 73-78. https://doi.org/10.1111/j.1365-3059.2006.01499.x
[4] Cheng et al. 2024, Nature 632, 823-831. https://doi.org/10.1038/s41586-024-07682-9
[5] Hafeez et al. 2025, Nature Plants. https://doi.org/10.1038/s41477-025-01920-2
[6] Brown et al. 2015, Fungal Genetics & Biology 79, 33-41. https://doi.org/10.1016/j.fgb.2015.04.017
[7] Nelson et al. 2018, Nature Reviews Genetics 19, 21-33. https://doi.org/10.1038/nrg.2017.82
[8] Battache et al. 2024, BMC Plant Biology. https://doi.org/10.1186/s12870-024-05426-5
Follow the Topic
-
Nature Plants

An online-only, monthly journal publishing the best research on plants — from their evolution, development, metabolism and environmental interactions to their societal significance.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in