Evolution of fast-growing piscivorous herring in the young Baltic Sea
Published in Ecology & Evolution and Genetics & Genomics
My studies of the population genetics of Atlantic herring started as a Bachelor of Science thesis project in the late 1970s! The project involved population samples from the species distribution in the Atlantic Ocean and the brackish Baltic Sea. This is before DNA-based methods had been developed and we used starch gel electrophoresis to detect genetic variation in the isoelectric point of various proteins. The project was successful in finding genetic variation and I could score genetic polymorphism at 13 loci from samples collected at 17 different localities, an impressive study at the time. However, I found the same alleles at the same frequencies in each population sample. We could not refute the null hypothesis that all herring belongs to a single panmictic population. This was an unexpected result, in particular since Linnaeus, the father of taxonomy and affiliated with Uppsala University like me, classified the small Baltic herring as a subspecies (Clupea harengus membras) of the much larger Atlantic herring (Clupe harengus harengus). After this “successful” thesis project, I left herring genetics and did my PhD on the genetics of domestic animals and continued to primarily work on population genomics of domestic animals for the next 30 years.
However, the herring results bothered me. How is it possible that a species reproducing under such variable environmental conditions do not show any genetic differentiation? One possible explanation was that there is essentially no genetic drift at neutral loci due to the huge population size of herring. If that is the case, the signals of genetic differentiation should be striking, due to the lack of noise caused by drift. So, when next generation sequencing became available, I decided to take up herring genetics, but now using whole genome sequencing of many population samples including some of the samples I collected as a student more than 40 years ago. The results have been amazing with hundreds of loci standing out as skyscrapers above a flat background when comparing allele frequencies in populations adapted to different environmental conditions. Four of these loci constitute megabase-sized inversions that were described in detail based on PacBio long-read sequencing in an article also published in Nature Communications during 2024.
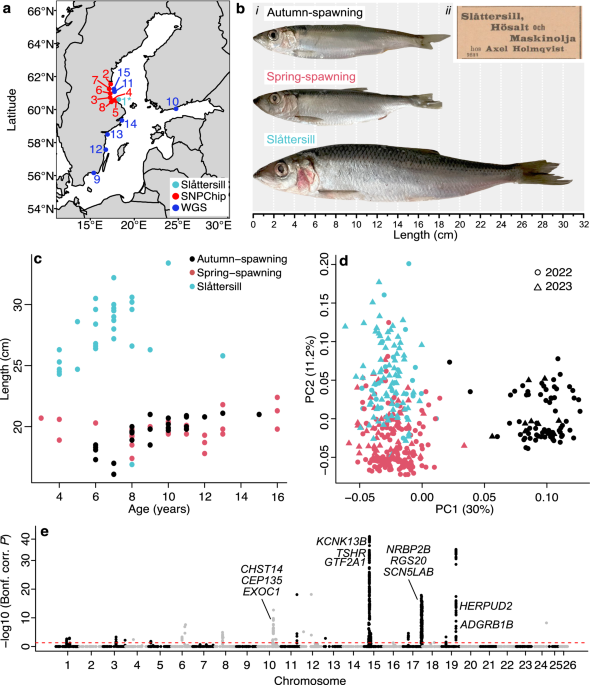
The current project was initiated after a discussion with a local fisherman who recommended us to sample a specific type of herring the locals call Slåttersill. These are named Slåttersill because they are spawning at hay harvesting time (slåtter in Swedish) and they are large as Atlantic herring (sill in Swedish), and thus much larger than the common Baltic herring. The fact that these fish are returning to the same area at the same time, year after year, raised my curiosity so we sampled the population at spawning and performed a genetic screen using a SNP-chip comprising 4,500 SNPs representing the majority of genomic loci showing genetic differentiation among populations. The result of the SNP screen immediately revealed that we had identified a genetically unique population.
The result prompted us to perform a detailed phenotypic characterisation which strongly indicated that Slåttersill have shifted to become piscivorous (fish-eating) in contrast to the planktivorous (plankton-eating) small-sized Baltic herring that by far is the dominating type in the Baltic Sea. This was indicated by a much faster growth-rate, higher fat content, lower level of dioxin contaminant all indicating a shift in diet and interestingly damaged gill rakers. A planktivorous fish, like the herring, has a dense set of gill rakers used for sieving plankton. The lesions observed in the piscivorous herring have most likely been caused by feeding on sticklebacks, a very abundant fish in the Baltic Sea with sharp spines for predation protection.
Next, we teamed up with researchers at the Swedish University of Agricultural Sciences that had a collection of exceptionally large herring collected from different parts of the Baltic Sea. Interestingly, the stomach content from these large herring showed that they had been feeding on small fish, and in particular sticklebacks. We next performed whole genome sequencing of all large herring and the results showed clear genetic differentiation from the small planktivorous Baltic herring and that there are at least two subpopulations of piscivorous herring in the Baltic Sea.
An interesting question is why fish-eating herring has evolved in the Baltic Sea, when there is no evidence for such herring in the Atlantic Ocean? The Baltic Sea is a young water body that has only existed for about 8,000 years after the end of the last glaciation period. Only a limited number of marine fish have been able to colonize the brackish Baltic Sea, where salinity is in the range 2-10‰ compared with about 35‰ in the Atlantic Ocean. Thus, fish-eating Baltic herring has most likely evolved due to lack of competition from other predatory fish, for instance, mackerel and tuna.
This study, as well as our previous studies on Atlantic and Baltic herring, show that there exists a considerable amount of unrecognized biodiversity within a superabundant species like the herring. This genetic diversity will likely be crucial for how the herring can adapt to future climate change and for future food security as herring constitutes about 70% of all fish caught in the Baltic Sea. The current stock management of herring in the Baltic Sea does not at all take into account the presence of many genetically distinct populations in this area, and some of these, like the fish-eating Baltic herring, are probably very sensitive to overfishing.
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in