The age that an animal reaches sexual or reproductive maturity is a key determinant of how quickly a population can grow, or bounce back after a period of decline. Because of this, age at maturity can be used as a proxy for species extinction risk, or be fed into more sophisticated models to understand population growth dynamics and species resilience. The problem is, getting age at maturity data in the field is notoriously difficult and time-consuming.
We have developed models that estimate age at maturity in vertebrates that requires only knowledge of a species’ genome sequence. Thus, the method is applicable to any vertebrate with a moderate-quality genome assembly and is both rapid and cost-effective. For comparison, field observations for age at maturity can take decades and cost upwards of $50,000 USD. Including genome sequencing, our method costs just $5000 and takes under two weeks, or $0 and one day if genome sequencing and assembly is already complete.

The challenge of managing endangered species is becoming increasingly urgent. More than 20 per cent of vertebrate species are under threat, and for many we lack fundamental data needed for their optimal management. This data includes species’ lifespan and age at maturity.
The age at which animals reach sexual or reproductive maturity profoundly affects their capacity for population growth because it is a direct indication of generation time and provides key information on reproductive output. Where age at maturity is known, it can form a foundation for species management strategies or provide crucial data for species resilience models.
Yet, measuring age at sexual maturity is difficult, especially for long-lived or elusive species. For fish, it often relies on dissecting individuals to view the developmental status of cells in the gonads. For mammals, it typically relies on observations and recognition of secondary sexual characteristics (e.g. antlers, tusks) or the presence of pregnant individuals in wild populations. In some marine mammals, age at maturity is even estimated using faeces steroid hormone levels.
Although existing methods are useful and often precise, they are generally very costly, time consuming and often species-specific. This limits not only their applicability but also prevents cross species comparisons. Given the urgency of this fundamental biological information for the assessment of threatened species, a more rapid, cost effective and universal approach is required.
Predicting age at maturity
Previous research by CSIRO, Australia’s national science agency, has shown the frequency of a distinctive pattern of DNA sequence known as “CpG sites” (i.e., CG sequences) in gene promoters can be used to predict lifespan in mammals, fish and other vertebrates. In the current paper, we asked whether promoter CpG content can also predict the age at which any vertebrate species reaches maturity.
To develop our predictive model, we compiled a dataset consisting of all vertebrate species with suitable, publicly available genome sequences and age at maturity data (n = 1359 species, including fish, birds and reptiles, mammals, and amphibians). We sourced genome sequences from NCBI and identified gene promoters using BLAST+ with Eukaryotic Promoter Database reference sequences.
Age at maturity data were source from several pre-existing databases, including AnAge, FishBase and directly from the literature. Known ages at maturity ranged from 0.06 ± 0.006 years in the California vole (Microtus californicus) to 25.52 ± 0.87 years in the Aldabra giant tortoise (Aldabrachelys gigantea).

We trained and tested our predictive model and evaluated improvements in prediction accuracy gained by developing group-specific models; one each for fish, mammals, and reptiles (including birds). Cross-validation demonstrated strong correlations between known and predicted ages, especially for mammals, and a median prediction error of about 30%, or just under one year.
We also took some additional steps to validate our predictions. Firstly, we generated prediction intervals for individual age at maturity estimates to allow for better interpretation of prediction uncertainty, which varies from species to species. Secondly, we predicted age at maturity for genome assemblies from multiple individuals of the same species. The results showed little variance between individuals, indicating that the genome sequence of a single individual can produce an age at maturity prediction that is representative for the entire species.
Predicted ages
We used our predictive model to estimate the age at maturity for 1912 species of unknown age at maturity, for which genome sequences were publicly available.
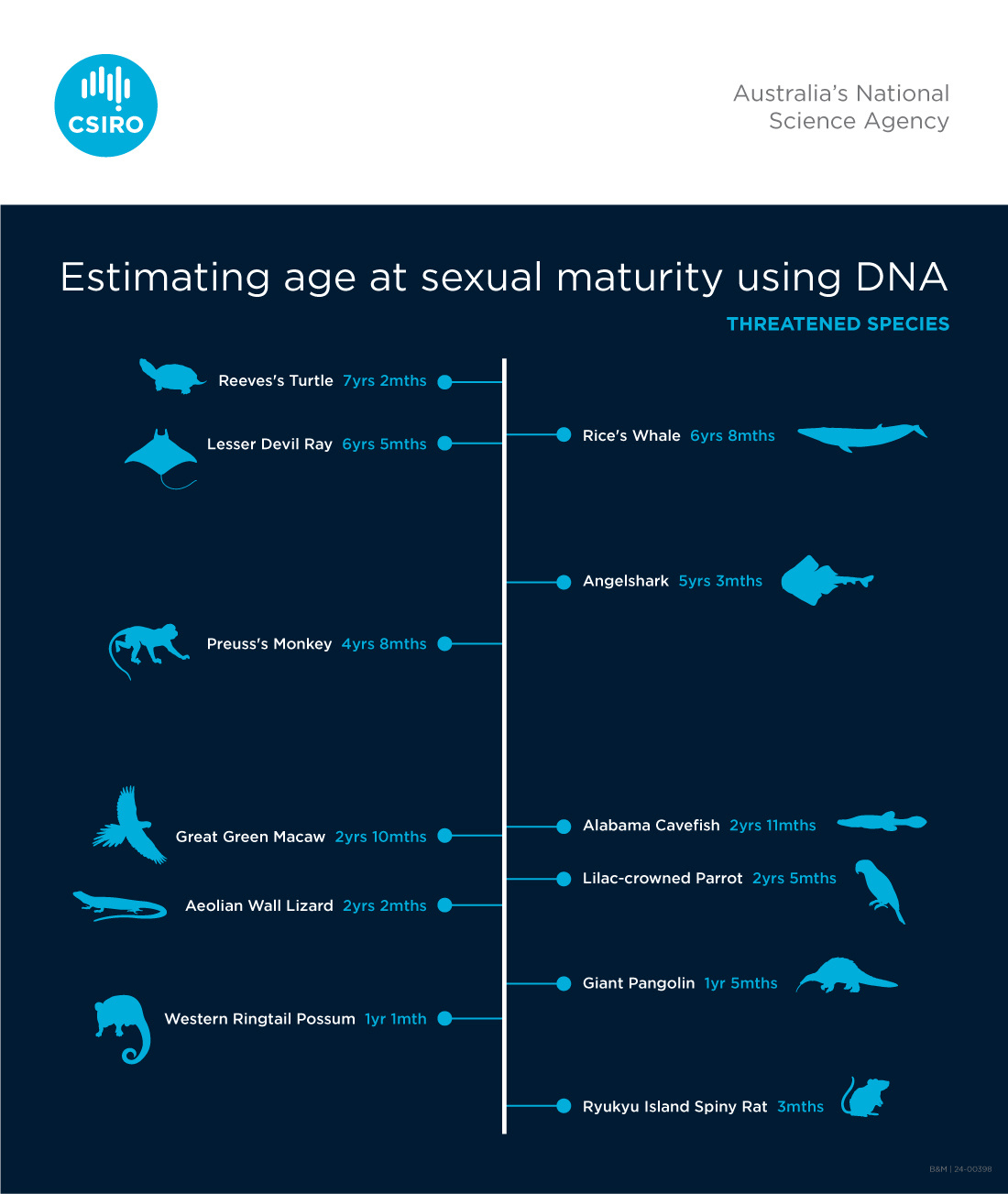
For a randomly selected subset of species, new age at maturity predictions were compared with known values for closely related species. This allowed us to provide an indication of probable accuracy in the absence of reported values for ground truthing. Predicted ages at maturity typically fell within the range of closely related species, giving us even greater confidence in the models.
Reflections
What excites us about this method is how broadly applicable it is. It only requires a moderate-quality genome assembly, making it accessible for many species. The results are especially relevant for species conservation and management, as knowing life history traits like generation time supports the assessment of extinction risk, as well as other measures like the risk of overfishing.
But, how does something as simple as gene promoter sequence content predict such fundamental biological traits as lifespan and in the case of our study, age at maturity? We don’t have a complete picture yet, but our model shows that genes whose promoters are most strongly associated with age at maturity are either expressed in reproductive tissues or have specific roles in reproductive maturation. This is a clue that the model is picking up on regulatory information specific to our trait of interest. This may be related to DNA methylation, a repressive mark that occurs most frequently at CpG sites, or other transcriptional changes associated with the promoter regions we've identified.
Genomic and epigenomic biomarkers present exciting, practical and transformative new ways to understand animal ecology. With methods to predict species age at maturity, species lifespan and individual age already developed, questions remain about how can these assays be optimised and what other important measures can epigenomics yield? As genome data becomes exponentially more available, we will have access to additional data to improve the accuracy of existing models, and provide opportunities to develop methods for non-vertebrate groups. We will be better positioned to develop new biomarkers for additional traits such as individual age at maturity, sex and even stress.
We anticipate that soon, efficient and cost-effective 'omics methods will routinely support biodiversity conservation by providing us access to the information we need to better manage species globally.
Follow the Topic
-
Communications Biology

An open access journal from Nature Portfolio publishing high-quality research, reviews and commentary in all areas of the biological sciences, representing significant advances and bringing new biological insight to a specialized area of research.
Related Collections
With Collections, you can get published faster and increase your visibility.
Signalling Pathways of Innate Immunity
Publishing Model: Hybrid
Deadline: Feb 28, 2026
Forces in Cell Biology
Publishing Model: Open Access
Deadline: Apr 30, 2026





Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in