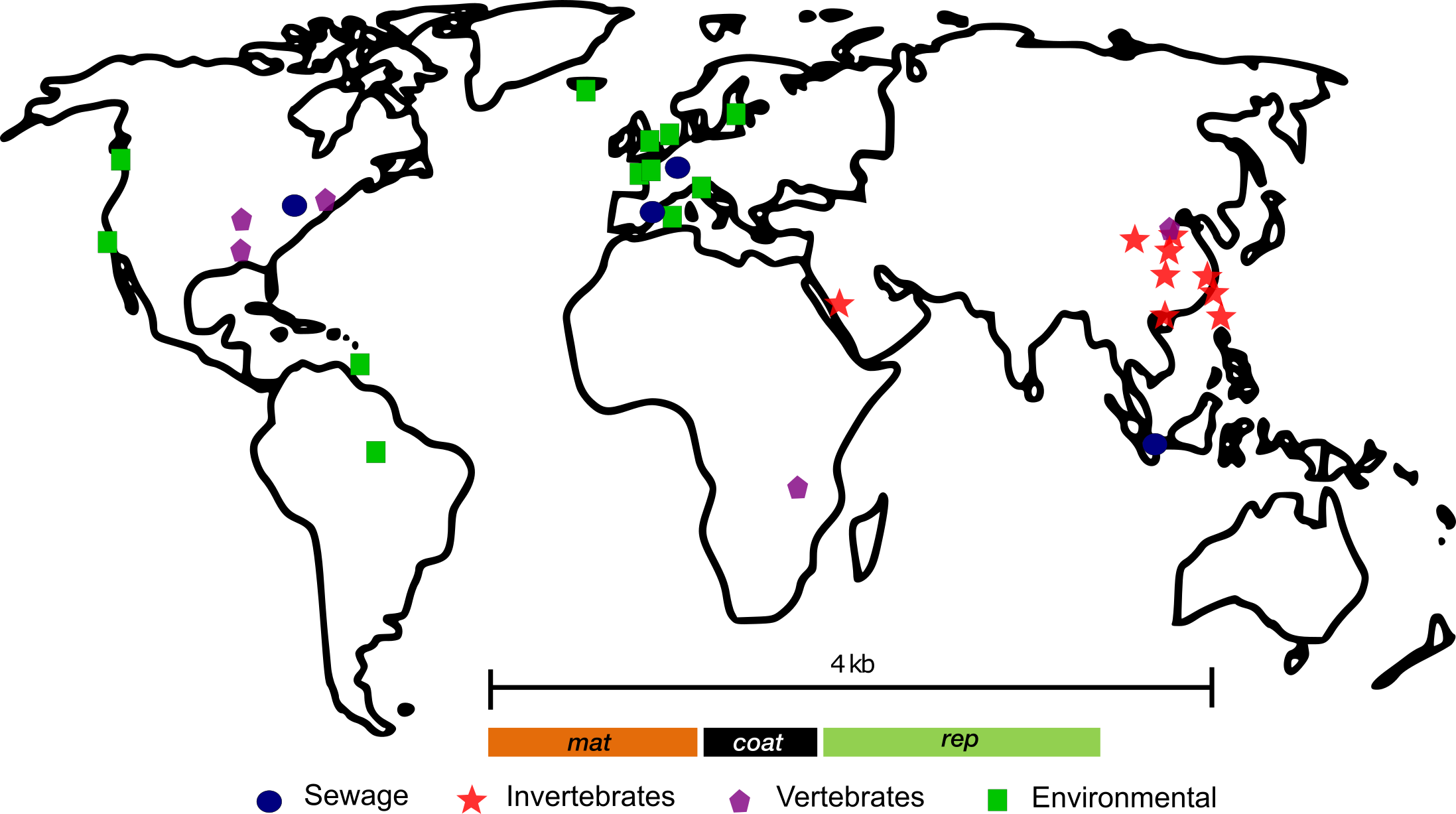
The source locations of each unique metatranscriptome are mapped on the world map and the type of the source material is indicated by different colored symbols. A generic ssRNA phage genome with three core genes (mat, coat, and rep) is shown as an inset.
Caudovirales, the big dsDNA phages that dominate the virosphere, encode multiple proteins for active destruction of the cell envelope, including at least one cell wall-degrading enzyme. In contrast, small ssDNA and ssRNA phages use a single, non-catalytic protein that induces the cell to self-destruct (“autolysis”). Moreover, because of the cramped genomic conditions (typically 3 - 5 kb), the “sgl” (Single Gene Lysis) gene is usually found embedded out of frame inside another essential phage gene. Considering that each Sgl is in effect pushing a “magic button” for bacteriolysis, our lab has been interested in determining each lytic mechanism. In studies with classical ssRNA and ssDNA phages, 10 Sgls have been identified. For three Sgls, we have shown that each binds to and inhibits a different enzyme of the conserved peptidoglycan (PG) synthesis pathway: proteins E, A2, and LysM of phages φX174 (ssDNA), Qβ (ssRNA) and M (ssRNA) inhibit MraY, MurA and MurJ, respectively. And, just as in treatment with beta-lactam cell wall antibiotics, lysis with these “PA” (protein antibiotic) Sgls involves a septal catastrophe when the intoxicated cell attempts to undergo cell division without PG biosynthesis. A fourth Sgl, the L protein of ssRNA phage MS2, is still unsolved in terms of its mechanism, although we know that it does not inhibit cell wall synthesis. Recently we have provided evidence that the five of the other six classic Sgls operate like L, although lacking significant sequence similarity. So, although we still had work to do, we were closing in on some degree of completion, confident that future Sgls would turn out to be either PA-type or L-like.
Then, in 2016, our comfortable little corner of the phage world just exploded, and has kept exploding ever since. Two papers reported the assembly of more than 200 complete ssRNA genomes, mostly from meta-transcriptome datasets1,2. Surprisingly, only one of these genomes had an identifiable sgl, which was found because it was ~38% identical to MS2 L. With only genomic information and no idea of the host, we couldn’t do classical genetics with these “metaphages”, within each of which we expected to find an sgl embedded in one of the three core genes of the ssRNA phage. Using reasonable bioinformatic constraints (Figure 2), we picked out candidate sgl open reading frames, had them synthesized, cloned them into an inducible expression plasmid, and tested them for lethal activity in Escherichia coli.
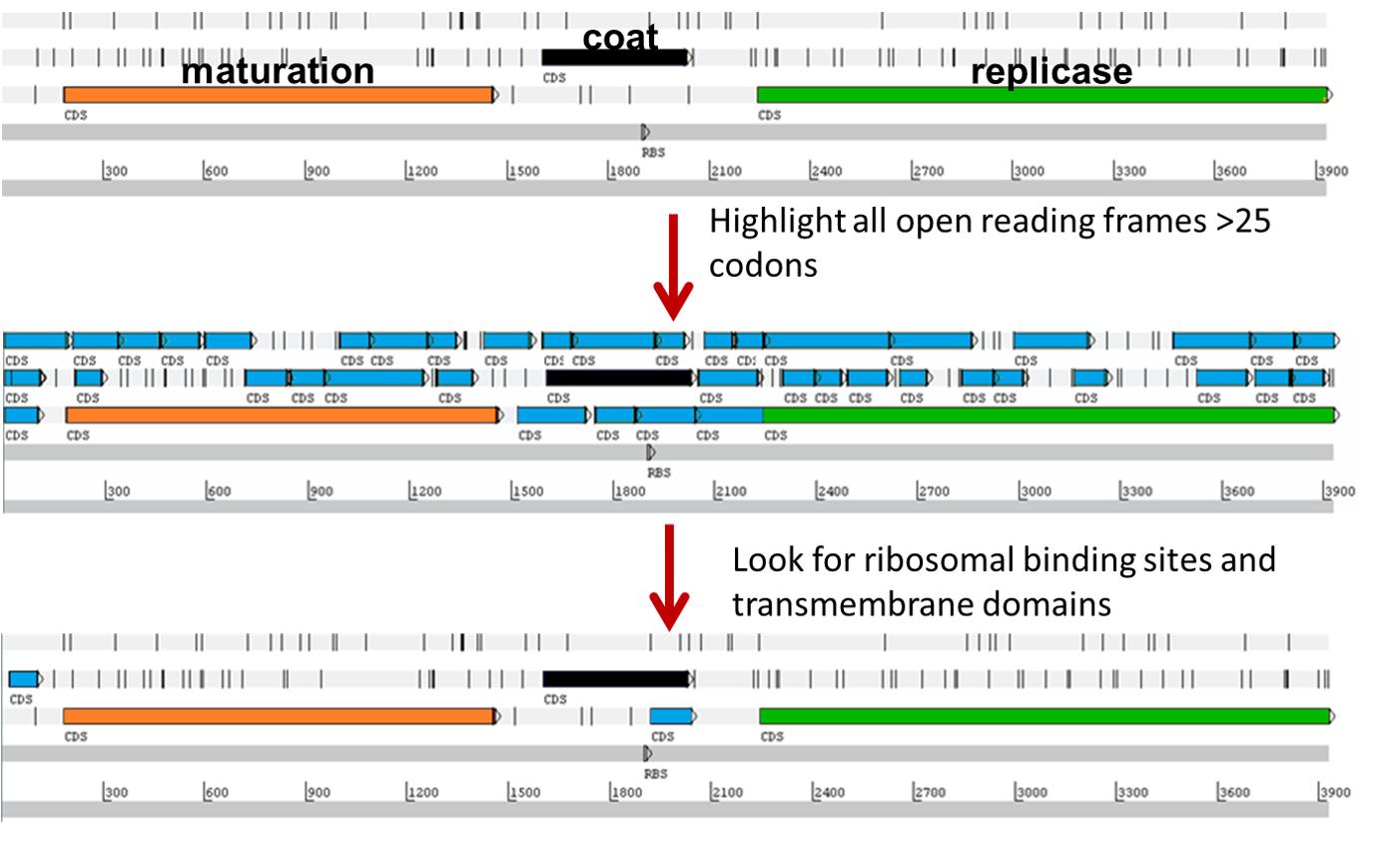
Of the 293 candidates tested, 33 were functional in the initial screen (Figure 3) and two more were later made functional by directed evolution. Interestingly, many genomes had more than one Sgl candidate. Since each ssRNA phage relies on a specific retractable pilus as its receptor, and since such pili are often encoded on mobile genetic elements, we reasoned that these phages often suddenly find themselves in a new host requiring a different Sgl for lysis. In support of this notion, of two candidate sgl genes in the genome Beihai9, one proved to be functional in E. coli and the other could be converted into lytic functionality by directed evolution, in this case requiring 2 single base changes. Considering the extremely high mutation rate in ssRNA phages (>10-4 per base per replication), this indicates that, despite the extremely limited genomic space, these phages can encode two or more Sgls or proto-Sgls and thus be prepared for lytic propagation in multiple hosts. Not surprisingly, further examination of genomes of closely related phages revealed abundant evidence of de novo gene evolution.
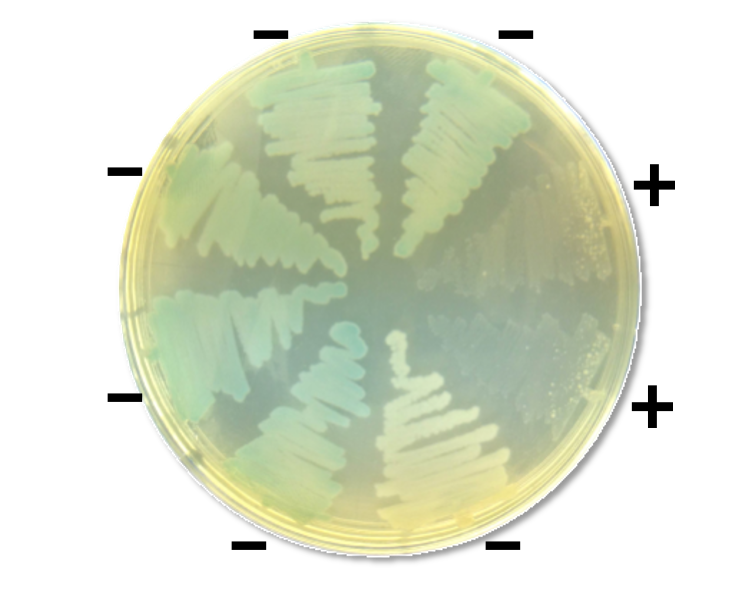
The sgl candidates were synthesized and cloned into arabinose inducible vector and screened for function. The sgl candidates that showed growth inhibition were scored as functional (+) and one with no discernible growth defect as non-functional (-).
As if the expansion from ~10 known Sgls to >200 was not enough, metatranscriptome analysis in the last year has seen two more logarithmic jumps in the number of new ssRNA genome, first ~1000 from soil samples3 and then ~16,000 more from a systematic analysis of publicly available datasets4. Not surprisingly, very few of the functional Sgls from our study had homologs in the new genomes. This suggests that, out there, ssRNA viral “dark matter” contains a mind-boggling diversity of Sgls, potentially representing every possible way of causing bacterial autolysis.
- Krishnamurthy, S. R., Janowski, A. B., Zhao, G., Barouch, D. & Wang, D. Hyperexpansion of RNA Bacteriophage Diversity. PLoS Biol 14, e1002409, doi:10.1371/journal.pbio.1002409 (2016).
- Shi, M. et al. Redefining the invertebrate RNA virosphere. Nature 540, 539-543, doi:10.1038/nature20167 (2016).
- Starr, E. P., Nuccio, E. E., Pett-Ridge, J., Banfield, J. F. & Firestone, M. K. Metatranscriptomic reconstruction reveals RNA viruses with the potential to shape carbon cycling in soil. Proc Natl Acad Sci U S A 116, 25900-25908, 518 doi:10.1073/pnas.1908291116 (2019).
- Callanan, J. et al. Expansion of known ssRNA phage genomes: From tens to over a thousand. Science advances 6, eaay5981, doi:10.1126/sciadv.aay5981 (2020).
Follow the Topic
-
Nature Communications

An open access, multidisciplinary journal dedicated to publishing high-quality research in all areas of the biological, health, physical, chemical and Earth sciences.
Related Collections
With Collections, you can get published faster and increase your visibility.
Women's Health
Publishing Model: Hybrid
Deadline: Ongoing
Advances in neurodegenerative diseases
Publishing Model: Hybrid
Deadline: Mar 24, 2026
Latest Content
Why is Singapore Identified in Global Research as Number One? How Physical Activity and Education Excellence Created a Global Leader






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in