Intestinal regeneration: lessons from organoids
Published in Bioengineering & Biotechnology

As a lab interested in tissue self-organization, we found small intestinal organoids to be a perfect model: they can develop from a single cell, driven by the intrinsic capacity of a cell to undergo a regenerative process, building a complicated hierarchical structure through self-organization from the bottom up. However, it was not previously known which factors drive and regulate this process, with many labs reporting the importance of various signaling pathways, but few systematic studies addressing the underlying complex networks of functional interactions. We set out to generate the first map of such interactions using a broad coverage annotated compound library for an image-based phenotypic screen, but quickly realized the associated challenges. Most organoid protocols rely on tedious manual steps and lack reproducibility and robustness. To perform the screen, we needed to make sure we could image a sufficient number of organoids that we first needed to culture, then fix and stain for cell type markers. A lack of out-of-the-box solutions meant we had to be resourceful: firstly, we had to miniaturize the assay and make it high throughput-compatible. Our final workflow was fully automated, had reproducible organoid growth and included only 2 medium exchange steps followed by an automated immunofluorescence sample handling pipeline to process and stain Matrigel-embedded organoids (Figure 1).
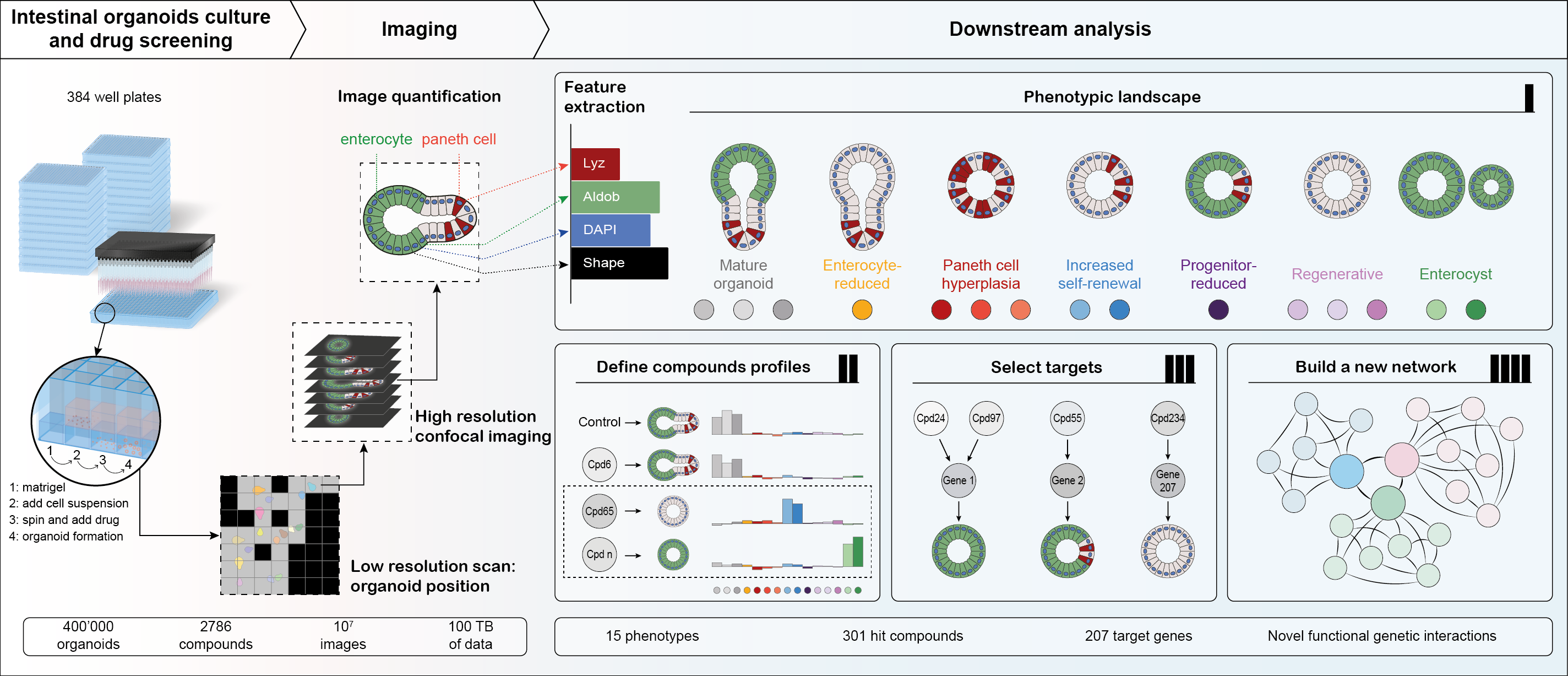
Screening workflow and analysis pipeline: Organoids were cultured, fixed stained and imaged in 384 well plates. Images were processed to segment organoids and extract features. Phenotypic landscape consisted of 15 phenotypes, producing condition-specific signatures that were used to select hits and infer interactions.
We soon realized that this was, in fact, the easy part: we now needed to develop an imaging pipeline to image hundreds of thousands of three-dimensional structures that can vary greatly in size and are distributed randomly over the entire well. We solved this with an iterative imaging workflow, first acquiring each well at low resolution, segmenting on-the-fly, and then re-acquiring only the detected objects with high magnification 3D confocal imaging. Although reducing the acquisition time and generated data amount by up to 60%, we nonetheless ended up with over 100TB of imaging data that enabled us to describe over 400,000 organoids at single-cell resolution, generating an unprecedented dataset.
The next task was similarly challenging: how do we determine the phenotypes, having no a priori assumption? We turned to a data-driven approach, using the multivariate feature space to infer 15 phenotypes. To our surprise, there was no single “wild-type” phenotype, but rather a distribution, reflecting the variability of the system. These distributions were, however, condition-specific, giving a unique phenotypic fingerprint that we used for two main purposes: defining the hits of the screen, but also inferring functional genetic interactions.
Our phenotypic fingerprint approach allowed us to find compounds with a regenerative phenotypic signature, one of which, an RXR antagonist, we profiled in-depth (Figure 2), making a number of striking discoveries:
- RXR inhibition results in YAP nuclear retention, leading to maintained regenerative phenotype. Interestingly, this inhibitor alone does not cause, by itself, an activation of Yap, it only maintains the activated state
- RXR-mediated signaling is required for enterocyte differentiation and exit from the regenerative state
- RA signaling in homeostasis is regulated by retinol metabolism and is crucial for acquiring intestinal identity after regeneration
- RXR antagonist treatment keeps the organoid in a fetal-like regenerative state
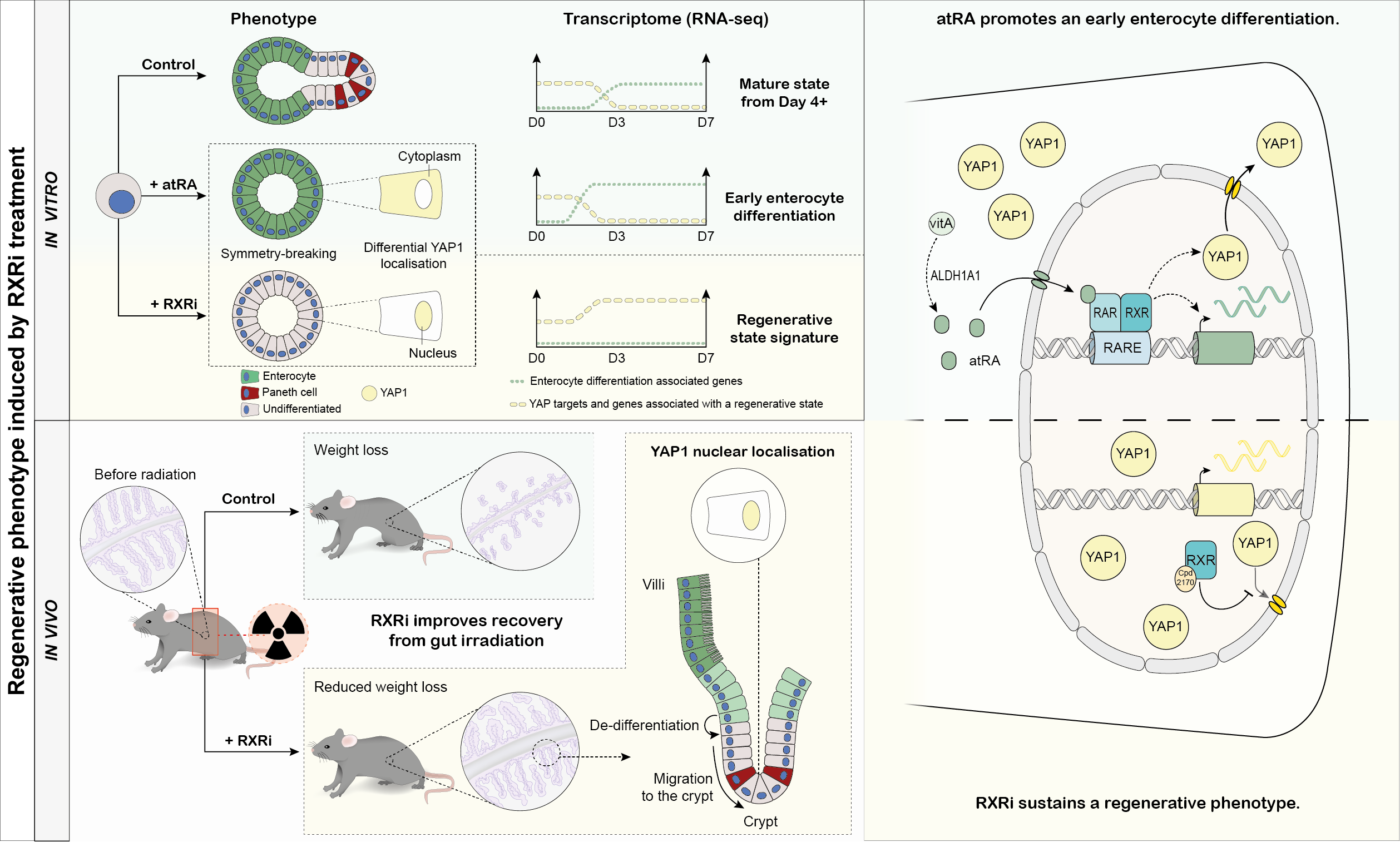
Validation of the compound effect in organoids and in vivo: RXRi-treated organoids retain early, regenerative transcriptome, whereas both intracellularly produced and exogenous retinoic acid promotes enterocyte maturation. In irradiated mice, RXRi treatment improves tissue regeneration and reduces weight loss.
Our data on role of retinoic acid metabolism proves it is a key factor in intestinal cell maturation, this insight could be instrumental to make sure iPSC-derived intestinal organoids manage to from mature tissue phenotypes. The role of RXR in regeneration has turned out to be even more fascinating: prompted by the organoid data, we investigated whether the regenerative phenotype in organoids translated into improved regeneration in vivo. For this, we teamed up with colleagues from the Genomics Institute of the Novartis Foundation (GNF). What we saw, was beyond our expectations: gamma-irradiated mice treated with RXR inhibitor recovered better from intestinal damage, showing improved tissue morphology and reduced weight loss. Combined with our discoveries in organoids, this meant we had found a compound that selectively affected regenerative cells, prolonging the time cells spend in a regenerative state without causing uncontrolled cell division. We believe that our finding paves the way for novel therapies to promote regeneration and recovery of the intestinal epithelium following acute damage, for instance in cancer patients receiving chemo- or radiation therapy.
This post was written by Ilya Lukonin and Prisca Liberali. Graphics by Veronique Juvin (sciartwork.com).
Follow the Topic
-
Nature

A weekly international journal publishing the finest peer-reviewed research in all fields of science and technology on the basis of its originality, importance, interdisciplinary interest, timeliness, accessibility, elegance and surprising conclusions.



Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in